Discovering Statistics Using SPSS
Andy Field (3rd Edition, 2009)
CONTENT
Chapter 1: Why is my evil lecturer forcing me to learn statistics?
Chapter 2: Everything you ever wanted to know about statistics
Chapter 4: Exploring data with graphs
Chapter 5: Exploring assumptions
Chapter 6: Correlation
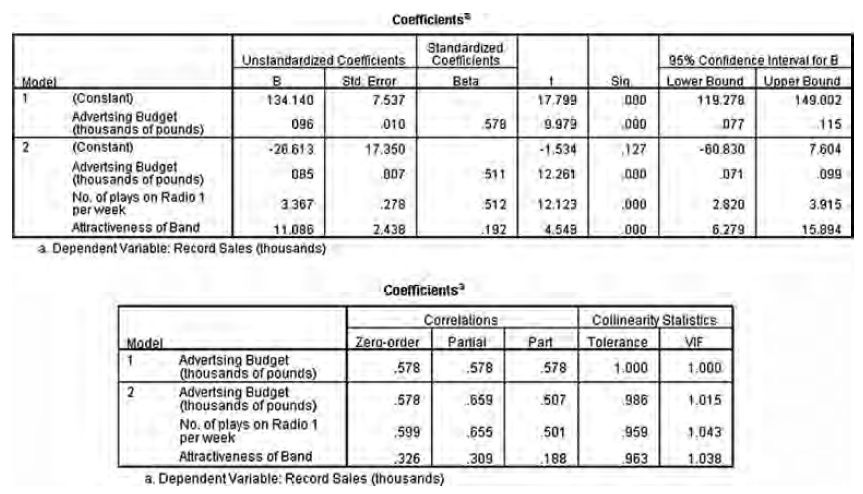
Chapter 7: Regression
Chapter 8: Logistic regression
Chapter 9: Comparing two means
Chapter 10: Comparing several means, ANOVA (GLM 1)
Chapter 11: Analysis of covariance, ANCOVA (GLM 2)
Chapter 13: Repeated-measures designs (GLM 4)
Chapter 15: Non-parametric tests
Chapter 17: Exploratory factor analysisChapter 1: Why is my evil lecturer forcing me to learn statistics?
A categorical variable is made up of categories. A categorical variable that you should be familiar with already is your species (e.g. human, domestic cat, fruit bat, etc.). You are a human or a cat or a fruit bat: you cannot be a bit of a cat and a bit of a bat, and neither a batman nor (despite many fantasies to the contrary) a catwoman (not even one in a nice PVC suit) exist. A categorical variable is one that names distinct entities. In its simplest form it names just two distinct types of things, for example male or female. This is known as a binary variable. Other examples of binary variables are being alive or dead, pregnant or not, and responding ‘yes’ or ‘no’ to a question. In all cases there are just two categories and an entity can be placed into only one of the two categories.
When two things that are equivalent in some sense are given the same name (or number), but there are more than two possibilities, the variable is said to be a nominal variable. It should be obvious that if the variable is made up of names it is pointless to do arithmetic on them (if you multiply a human by a cat, you do not get a hat). However, sometimes numbers are used to denote categories. For example, the numbers worn by players in a rugby or football (soccer) team. In rugby, the numbers of shirts denote specific field positions, so the number 10 is always worn by the fly-half (e.g. England’s Jonny Wilkinson), [8] and the number 1 is always the hooker (the ugly-looking player at the front of the scrum). These numbers do not tell us anything other than what position the player plays. We could equally have shirts with FH and H instead of 10 and 1. A number 10 player is not necessarily better than a number 1 (most managers would not want their fly-half stuck in the front of the scrum!). It is equally as daft to try to do arithmetic with nominal scales where the categories are denoted by numbers: the number 10 takes penalty kicks, and if the England coach found that Jonny Wilkinson (his number 10) was injured he would not get his number 4 to give number 6 a piggyback and then take the kick. The only way that nominal data can be used is to consider frequencies. For example, we could look at how frequently number 10s score tries compared to number 4s.
So far the categorical variables we have considered have been unordered (e.g. different brands of Coke with which you’re trying to kill sperm) but they can be ordered too (e.g. increasing concentrations of Coke with which you’re trying to kill sperm). When categories are ordered, the variable is known as an ordinal variable. Ordinal data tell us not only that things have occurred, but also the order in which they occurred. However, these data tell us nothing about the differences between values. Imagine we went to a beauty pageant in which the three winners were Billie, Freema and Elizabeth. The names of the winners don’t provide any information about where they came in the contest; however labelling them according to their performance does – first, second and third. These categories are ordered. In using ordered categories we now know that the woman who won was better than the women who came second and third. We still know nothing about the differences between categories, though. We don’t, for example, know how much better the winner was than the runners-up: Billie might have been an easy victor, getting much higher ratings from the judges than Freema and Elizabeth, or it might have been a very close contest that she won by only a point. Ordinal data, therefore, tell us more than nominal data (they tell us the order in which things happened) but they still do not tell us about the differences between points on a scale.
The next level of measurement moves us away from categorical variables and into continuous variables. A continuous variable is one that gives us a score for each person and can take on any value on the measurement scale that we are using. The first type of continuous variable that you might encounter is an interval variable. Interval data are considerably more useful than ordinal data and most of the statistical tests in this book rely on having data measured at this level. To say that data are interval, we must be certain that equal intervals on the scale represent equal differences in the property being measured. For example, on http://www.ratemyprofessors.com students are encouraged to rate their lecturers on several dimensions (some of the lecturers’ rebuttals of their negative evaluations are worth a look). Each dimension (i.e. helpfulness, clarity, etc.) is evaluated using a 5-point scale. For this scale to be interval it must be the case that the difference between helpfulness ratings of 1 and 2 is the same as the difference between say 3 and 4, or 4 and 5. Similarly, the difference in helpfulness between ratings of 1 and 3 should be identical to the difference between ratings of 3 and 5. Variables like this that look interval (and are treated as interval) are often ordinal – see Jane Superbrain Box 1.2.
1.7. Analysing data
Once you’ve collected some data a very useful thing to do is to plot a graph of how many times each score occurs. This is known as a frequency distribution, or histogram, which is a graph plotting values of observations on the horizontal axis, with a bar showing how many times each value occurred in the data set. Frequency distributions can be very useful for assessing properties of the distribution of scores. We will find out how to create these types of charts in Chapter 4.
Frequency distributions come in many different shapes and sizes. It is quite important, therefore, to have some general descriptions for common types of distributions. In an ideal world our data would be distributed symmetrically around the centre of all scores. As such, if we drew a vertical line through the centre of the distribution then it should look the same on both sides. This is known as a normal distribution and is characterized by the bell-shaped curve with which you might already be familiar. This shape basically implies that the majority of scores lie around the centre of the distribution (so the largest bars on the histogram are all around the central value).
Also, as we get further away from the centre the bars get smaller, implying that as scores start to deviate from the centre their frequency is decreasing. As we move still further away from the centre our scores become very infrequent (the bars are very short). Many naturally occurring things have this shape of distribution. For example, most men in the UK are about 175 cm tall; [12] some are a bit taller or shorter but most cluster around this value. There will be very few men who are really tall (i.e. above 205 cm) or really short (i.e. under 145 cm). An example of a normal distribution is shown in Figure 1.3.
There are two main ways in which a distribution can deviate from normal: (1) lack of symmetry (called skew) and (2) pointyness (called kurtosis). Skewed distributions are not symmetrical and instead the most frequent scores (the tall bars on the graph) are clustered at one end of the scale. So, the typical pattern is a cluster of frequent scores at one end of the scale and the frequency of scores tailing off towards the other end of the scale. A skewed distribution can be either positively skewed (the frequent scores are clustered at the lower end and the tail points towards the higher or more positive scores) or negatively skewed (the frequent scores are clustered at the higher end and the tail points towards the lower or more negative scores). Figure 1.4 shows examples of these distributions.
Distributions also vary in their kurtosis. Kurtosis, despite sounding like some kind of exotic disease, refers to the degree to which scores cluster at the ends of the distribution (known as the tails) and how pointy a distribution is (but there are other factors that can affect how pointy the distribution looks – see Jane Superbrain Box 2.2). A distribution with positive kurtosis has many scores in the tails (a so-called heavy-tailed distribution) and is pointy. This is known as a leptokurtic distribution. In contrast, a distribution with negative kurtosis is relatively thin in the tails (has light tails) and tends to be flatter than normal. This distribution is called platykurtic. Ideally, we want our data to be normally distributed (i.e. not too skewed, and not too many or too few scores at the extremes!). For everything there is to know about kurtosis read DeCarlo (1997).
In a normal distribution the values of skew and kurtosis are 0 (i.e. the tails of the distribution are as they should be). If a distribution has values of skew or kurtosis above or below 0 then this indicates a deviation from normal: Figure 1.5 shows distributions with kurtosis values of +1 (left panel) and –4 (right panel).
Chapter 2: Everything you ever wanted to know about statistics (well, sort of)
JANE SUPERBRAIN 2.2 Degrees of freedom
Degrees of freedom (df) is a very difficult concept to explain. I’ll begin with an analogy. Imagine you’re the manager of a rugby team and you have a team sheet with 15 empty slots relating to the positions on the playing field. There is a standard formation in rugby and so each team has 15 specific positions that must be held constant for the game to be played. When the first player arrives, you have the choice of 15 positions in which to place this player. You place his name in one of the slots and allocate him to a position (e.g. scrum-half) and, therefore, one position on the pitch is now occupied. When the next player arrives, you have the choice of 14 positions but you still have the freedom to choose which position this player is allocated. However, as more players arrive, you will reach the point at which 14 positions have been filled and the final player arrives. With this player you have no freedom to choose where they play – there is only one position left. Therefore there are 14 degrees of freedom; that is, for 14 players you have some degree of choice over where they play, but for 1 player you have no choice. The degrees of freedom is one less than the number of players.
In statistical terms the degrees of freedom relate to the number of observations that are free to vary. If we take a sample of four observations from a population, then these four scores are free to vary in any way (they can be any value). However, if we then use this sample of four observations to calculate the standard deviation of the population, we have to use the mean of the sample as an estimate of the population’s mean. Thus we hold one parameter constant. Say that the mean of the sample was 10; then we assume that the population mean is 10 also and we keep this value constant. With this parameter fixed, can all four scores from our sample vary? The answer is no, because to keep the mean constant only three values are free to vary. For example, if the values in the sample were 8, 9, 11, 12 (mean = 10) and we changed three of these values to 7, 15 and 8, then the final value must be 10 to keep the mean constant. Therefore, if we hold one parameter constant then the degrees of freedom must be one less than the sample size. This fact explains why when we use a sample to estimate the standard deviation of a population, we have to divide the sums of squares by N − 1 rather than N alone.
2.5. Going beyond the data
2.5.1. The standard error
We’ve seen that the standard deviation tells us something about how well the mean represents the sample data, but I mentioned earlier on that usually we collect data from samples because we don’t have access to the entire population. If you take several samples from a population, then these samples will differ slightly; therefore, it’s also important to know how well a particular sample represents the population. This is where we use the standard error. Many students get confused about the difference between the standard deviation and the standard error (usually because the difference is never explained clearly). However, the standard error is an important concept to grasp, so I’ll do my best to explain it to you.
We have already learnt that social scientists use samples as a way of estimating the behaviour in a population. Imagine that we were interested in the ratings of all lecturers (so, lecturers in general were the population). We could take a sample from this population. When someone takes a sample from a population, they are taking one of many possible samples. If we were to take several samples from the same population, then each sample has its own mean, and some of these sample means will be different.
Figure 2.7 illustrates the process of taking samples from a population. Imagine that we could get ratings of all lecturers on the planet and that, on average, the rating is 3 (this is the population mean, μ). Of course, we can’t collect ratings of all lecturers, so we use a sample. For each of these samples we can calculate the average, or sample mean. Let’s imagine we took nine different samples (as in the diagram); you can see that some of the samples have the same mean as the population but some have different means: the first sample of lecturers were rated, on average, as 3, but the second sample were, on average, rated as only 2. This illustrates sampling variation: that is, samples will vary because they contain different members of the population; a sample that by chance includes some very good lecturers will have a higher average than a sample that, by chance, includes some awful lecturers. We can actually plot the sample means as a frequency distribution, or histogram, [4] just like I have done in the diagram. This distribution shows that there were three samples that had a mean of 3, means of 2 and 4 occurred in two samples each, and means of 1 and 5 occurred in only one sample each. The end result is a nice symmetrical distribution known as a sampling distribution. A sampling distribution is simply the frequency distribution of sample means from the same population. In theory you need to imagine that we’re taking hundreds or thousands of samples to construct a sampling distribution, but I’m just using nine to keep the diagram simple. [5] The sampling distribution tells us about the behaviour of samples from the population, and you’ll notice that it is centred at the same value as the mean of the population (i.e. 3). This means that if we took the average of all sample means we’d get the value of the population mean. Now, if the average of the sample means is the same value as the population mean, then if we know the accuracy of that average we’d know something about how likely it is that a given sample is representative of the population. So how do we determine the accuracy of the population mean?
Think back to the discussion of the standard deviation. We used the standard deviation as a measure of how representative the mean was of the observed data. Small standard deviations represented a scenario in which most data points were close to the mean, a large standard deviation represented a situation in which data points were widely spread from the mean. If you were to calculate the standard deviation between sample means then this too would give you a measure of how much variability there was between the means of different samples. The standard deviation of sample means is known as the standard error of the mean (SE). Therefore, the standard error could be calculated by taking the difference between each sample mean and the overall mean, squaring these differences, adding them up, and then dividing by the number of samples. Finally, the square root of this value would need to be taken to get the standard deviation of sample means, the standard error.
Of course, in reality we cannot collect hundreds of samples and so we rely on approximations of the standard error. Luckily for us some exceptionally clever statisticians have demonstrated that as samples get large (usually defined as greater than 30), the sampling distribution has a normal distribution with a mean equal to the population mean, and a standard deviation of:
σX̄= s / √N (2.5)
This is known as the central limit theorem and it is useful in this context because it means that if our sample is large we can use the above equation to approximate the standard error (because, remember, it is the standard deviation of the sampling distribution). [6] When the sample is relatively small (fewer than 30) the sampling distribution has a different shape, known as a t-distribution, which we’ll come back to later.
2.6.2. One- and two-tailed tests
We saw in section 1.7.5 that hypotheses can be directional (e.g. ‘the more someone reads this book, the more they want to kill its author’) or non-directional (i.e. ‘reading more of this book could increase or decrease the reader’s desire to kill its author’). A statistical model that tests a directional hypothesis is called a one-tailed test, whereas one testing a non-directional hypothesis is known as a two-tailed test.
Imagine we wanted to discover whether reading this book increased or decreased the desire to kill me. We could do this either (experimentally) by taking two groups, one who had read this book and one who hadn’t, or (correlationally) by measuring the amount of this book that had been read and the corresponding desire to kill me. If we have no directional hypothesis then there are three possibilities. (1) People who read this book want to kill me more than those who don’t so the difference (the mean for those reading the book minus the mean for non-readers) is positive. Correlationally, the more of the book you read, the more you want to kill me – a positive relationship. (2) People who read this book want to kill me less than those who don’t so the difference (the mean for those reading the book minus the mean for non-readers) is negative. Correlationally, the more of the book you read, the less you want to kill me – a negative relationship. (3) There is no difference between readers and non-readers in their desire to kill me – the mean for readers minus the mean for non-readers is exactly zero. Correlationally, there is no relationship between reading this book and wanting to kill me. This final option is the null hypothesis. The direction of the test statistic (i.e. whether it is positive or negative) depends on whether the difference is positive or negative. Assuming there is a positive difference or relationship (reading this book makes you want to kill me), then to detect this difference we have to take account of the fact that the mean for readers is bigger than for non-readers (and so derive a positive test statistic). However, if we’ve predicted incorrectly and actually reading this book makes readers want to kill me less then the test statistic will actually be negative.
What are the consequences of this? Well, if at the .05 level we needed to get a test statistic bigger than say 10 and the one we get is actually -12, then we would reject the hypothesis even though a difference does exist. To avoid this we can look at both ends (or tails) of the distribution of possible test statistics. This means we will catch both positive and negative test statistics. However, doing this has a price because to keep our criterion probability of .05 we have to split this probability across the two tails: so we have .025 at the positive end of the distribution and .025 at the negative end. Figure 2.10 shows this situation – the tinted areas are the areas above the test statistic needed at a .025 level of significance. Combine the probabilities (i.e. add the two tinted areas together) at both ends and we get .05, our criterion value. Now if we have made a prediction, then we put all our eggs in one basket and look only at one end of the distribution (either the positive or the negative end depending on the direction of the prediction we make). So, in Figure 2.10, rather than having two small tinted areas at either end of the distribution that show the significant values, we have a bigger area (the lined area) at only one end of the distribution that shows significant values. Consequently, we can just look for the value of the test statistic that would occur by chance with a probability of .05. In Figure 2.10, the lined area is the area above the positive test statistic needed at a .05 level of significance. Note on the graph that the value that begins the area for the .05 level of significance (the lined area) is smaller than the value that begins the area for the .025 level of significance (the tinted area). This means that if we make a specific prediction then we need a smaller test statistic to find a significant result (because we are looking in only one tail of the distribution), but if our prediction happens to be in the wrong direction then we’ll miss out on detecting the effect that does exist. In this context it’s important to remember what I said in Jane Superbrain Box 2.4: you can’t place a bet or change your bet when the tournament is over. If you didn’t make a prediction of direction before you collected the data, youare too late to predict the direction and claim the advantages of a one-tailed test.
2.6.4. Effect sizes
The framework for testing whether effects are genuine that I’ve just presented has a few problems, most of which have been briefly explained in Jane Superbrain Box 2.6. The first problem we encountered was knowing how important an effect is: just because a test statistic is significant doesn’t mean that the effect it measures is meaningful or important. The solution to this criticism is to measure the size of the effect that we’re testing in a standardized way. When we measure the size of an effect (be that an experimental manipulation or the strength of a relationship between variables) it is known as an effect size. An effect size is simply an objective and (usually) standardized measure of the magnitude of observed effect. The fact that the measure is standardized just means that we can compare effect sizes across different studies that have measured different variables, or have used different scales of measurement (so an effect size based on speed in milliseconds could be compared to an effect size based on heart rates). Such is the utility of effect size estimates that the American Psychological Association is now recommending that all psychologists report these effect sizes in the results of any published work. So, it’s a habit well worth getting into.
Many measures of effect size have been proposed, the most common of which are Cohen’s d, Pearson’s correlation coefficient r (Chapter 6) and the odds ratio (Chapter 18). Many of you will be familiar with the correlation coefficient as a measure of the strength of relationship between two variables (see Chapter 6 if you’re not); however, it is also a very versatile measure of the strength of an experimental effect. It’s a bit difficult to reconcile how the humble correlation coefficient can also be used in this way; however, this is only because students are typically taught about it within the context of non-experimental research. I don’t want to get into it now, but as you read through Chapters 6, 9 and 10 it will (I hope) become clear what I mean. Personally, I prefer Pearson’s correlation coefficient, r, as an effect size measure because it is constrained to lie between 0 (no effect) and 1 (a perfect effect). [11] However, there are situations in which d may be favoured; for example, when group sizes are very discrepant r can be quite biased compared to d (McGrath & Meyer, 2006).
Effect sizes are useful because they provide an objective measure of the importance of an effect. So, it doesn’t matter what effect you’re looking for, what variables have been measured, or how those variables have been measured – we know that a correlation coefficient of 0 means there is no effect, and a value of 1 means that there is a perfect effect. Cohen (1988, 1992) has also made some widely used suggestions about what constitutes a large or small effect:
r = .10 (small effect): In this case the effect explains 1% of the total variance.
r = .30 (medium effect): The effect accounts for 9% of the total variance.
r = .50 (large effect): The effect accounts for 25% of the variance.
It’s worth bearing in mind that r is not measured on a linear scale so an effect with r = .6 isn’t twice as big as one with r = .3. Although these guidelines can be a useful rule of thumb to assess the importance of an effect (regardless of the significance of the test statistic), it is worth remembering that these ‘canned’ effect sizes are no substitute for evaluating an effect size within the context of the research domain that it is being used (Baguley, 2004; Lenth, 2001).
A final thing to mention is that when we calculate effect sizes we calculate them for a given sample. When we looked at means in a sample we saw that we used them to draw inferences about the mean of the entire population (which is the value in which we’re actually interested). The same is true of effect sizes: the size of the effect in the population is the value in which we’re interested, but because we don’t have access to this value, we use the effect size in the sample to estimate the likely size of the effect in the population. We can also combine effect sizes from different studies researching the same question to get better estimates of the population effect sizes. This is called meta-analysis – see Field (2001, 2005b).
2.6.5. Statistical power
Effect sizes are an invaluable way to express the importance of a research finding. The effect size in a population is intrinsically linked to three other statistical properties: (1) the sample size on which the sample effect size is based; (2) the probability level at which we will accept an effect as being statistically significant (the α-level); and (3) the ability of a test to detect an effect of that size (known as the statistical power, not to be confused with statistical powder, which is an illegal substance that makes you understand statistics better). As such, once we know three of these properties, then we can always calculate the remaining one. It will also depend on whether the test is a one- or two-tailed test (see section 2.6.2). Typically, in psychology we use an α-level of .05 (see earlier) so we know this value already. The power of a test is the probability that a given test will find an effect assuming that one exists in the population. If you think back you might recall that we’ve already come across the probability of failing to detect an effect when one genuinely exists (β, the probability of a Type II error). It follows that the probability of detecting an effect if one exists must be the opposite of the probability of not detecting that effect (i.e. 1 – β). I’ve also mentioned that Cohen (1988, 1992) suggests that we would hope to have a .2 probability of failing to detect a genuine effect, and so the corresponding level of power that he recommended was 1 – .2, or .8. We should aim to achieve a power of .8, or an 80% chance of detecting an effect if one genuinely exists. The effect size in the population can be estimated from the effect size in the sample, and the sample size is determined by the experimenter anyway so that value is easy to calculate. Now, there are two useful things we can do knowing that these four variables are related:
1 Calculate the power of a test: Given that we’ve conducted our experiment, we will have already selected a value of α, we can estimate the effect size based on our sample, and we will know how many participants we used. Therefore, we can use these values to calculate β, the power of our test. If this value turns out to be .8 or more we can be confident that we achieved sufficient power to detect any effects that might have existed, but if the resulting value is less, then we might want to replicate the experiment using more participants to increase the power.
2 Calculate the sample size necessary to achieve a given level of power: Given that we know the value of α and β, we can use past research to estimate the size of effect that we would hope to detect in an experiment. Even if no one had previously done the exact experiment that we intend to do, we can still estimate the likely effect size based on similar experiments. We can use this estimated effect size to calculate how many participants we would need to detect that effect (based on the values of α and β that we’ve chosen).
The latter use is the more common: to determine how many participants should be used to achieve the desired level of power. The actual computations are very cumbersome, but fortunately there are now computer programs available that will do them for you (one example is G*Power, which is free and can be downloaded from a link on the companion website; another is nQuery Adviser but this has to be bought!). Also, Cohen (1988) provides extensive tables for calculating the number of participants for a given level of power (and vice versa). Based on Cohen (1992) we can use the following guidelines: if we take the standard α-level of .05 and require the recommended power of .8, then we need 783 participants to detect a small effect size (r = .1), 85 participants to detect a medium effect size (r = .3) and 28 participants to detect a large effect size (r = .5).
Chapter 4: Exploring data with graphs
4.4. Histograms: a good way to spot obvious problems
The resulting histogram is shown in Figure 4.10. The first thing that should leap out at you is that there appears to be one case that is very different to the others. All of the scores appear to be squashed up at one end of the distribution because they are all less than 5 (yielding a very pointy distribution!) except for one, which has a value of 20! This is an outlier: a score very different to the rest (Jane Superbrain Box 4.1). Outliers bias the mean and inflate the standard deviation (you should have discovered this from the self-test tasks in Chapters 1 and 2) and screening data is an important way to detect them. You can look for outliers in two ways: (1) graph the data with a histogram (as we have done here) or a boxplot (as we will do in the next section), or (2) look at z-scores (this is quite complicated but if you want to know see Jane Superbrain Box 4.2).
The outlier shown on the histogram is particularly odd because it has a score of 20, which is above the top of our scale (remember, our hygiene scale ranged only from 0 to 4), and so it must be a mistake (or the person had obsessive compulsive disorder and had washed themselves into a state of extreme cleanliness). However, with 810 cases, how on earth do we find out which case it was? You could just look through the data, but that would certainly give you a headache and so instead we can use a boxplot which is another very useful graph for spotting outliers.
4.5. Boxplots (box–whisker diagrams)
Boxplots show us the range of scores, the range between which the middle 50% of scores fall, and the median, the upper quartile and lower quartile score. Like histograms, they also tell us whether the distribution is symmetrical or skewed. If the whiskers are the same length then the distribution is symmetrical (the range of the top and bottom 25% of scores is the same); however, if the top or bottom whisker is much longer than the opposite whisker then the distribution is asymmetrical (the range of the top and bottom 25% of scores is different). Finally, you’ll notice some circles above the male boxplot. These are cases that are deemed to be outliers. Each circle has a number next to it that tells us in which row of the data editor to find that case. In Chapter 5 we’ll see what can be done about these outliers.
JANE SUPERBRAIN 4.1 What is an outlier?
An outlier is a score very different from the rest of the data. When we analyse data we have to be aware of such values because they bias the model we fit to the data. A good example of this bias can be seen by looking at the mean. When the first edition of this book came out in 2000, I was quite young and became very excited about obsessively checking the book’s ratings on Amazon.co.uk. These ratings can range from 1 to 5 stars. Back in 2002, the first edition of this book had seven ratings (in the order given) of 2, 5, 4, 5, 5, 5, 5. All but one of these ratings are fairly similar (mainly 5 and 4) but the first rating was quite different from the rest – it was a rating of 2 (a mean and horrible rating). The graph plots seven reviewers on the horizontal axis and their ratings on the vertical axis and there is also a horizontal line that represents the mean rating (4.43 as it happens). It should be clear that all of the scores except one lie close to this line. The score of 2 is very different and lies some way below the mean. This score is an example of an outlier – a weird and unusual person (sorry, I mean score) that deviates from the rest of humanity (I mean, data set). The dashed horizontal line represents the mean of the scores when the outlier is not included (4.83). This line is higher than the original mean indicating that by ignoring this score the mean increases (it increases by 0.4). This example shows how a single score, from some mean-spirited badger turd, can bias the mean; in this case the first rating (of 2) drags the average down. In practical terms this had a bigger implication because Amazon rounded off to half numbers, so that single score made a difference between the average rating reported by Amazon as a generally glowing 5 stars and the less impressive 4.5 stars. (Nowadays Amazon sensibly produces histograms of the ratings and has a better rounding system.) Although I am consumed with bitterness about this whole affair, it has at least given me a great example of an outlier! (Data for this example were taken from http://www.amazon.co.uk/ in about 2002.)
JANE SUPERBRAIN 4.2 Using z-scores to find outliers
To check for outliers we can look at z-scores. We saw in section 1.7.4 that z-scores are simply a way of standardizing a data set by expressing the scores in terms of a distribution with a mean of 0 and a standard deviation of 1. In doing so we can use benchmarks that we can apply to any data set (regardless of what its original mean and standard deviation were). We also saw in this section that to convert a score into z-scores we simply take each score (X) and convert it to a z-score by subtracting the mean of all scores (X-bar) from it and dividing by the standard deviation of all scores (s).
We can get SPSS to do this conversion for us using the ANALYZE, DESCRIPTIVE STATISTICS, DESCRIPTIVES… dialog box, selecting a variable (such as day 2 of the hygiene data as in the diagram), or several variables, and selecting the Save standardized values as variables before we click on OK. SPSS creates a new variable in the data editor (with the same name prefixed with the letter z). To look for outliers we could use these z-scores and count how many fall within certain important limits. If we take the absolute value (i.e. we ignore whether the z-score is positive or negative) then in a normal distribution we’d expect about 5% to have absolute values greater than 1.96 (we often use 2 for convenience), and 1% to have absolute values greater than 2.58, and none to be greater than about 3.29.
Alternatively, you could use some SPSS syntax in a syntax window to create the z-scores and count them for you. I’ve written the file Outliers (Percentage of Z-scores). sps (on the companion website) to produce a table for day 2 of the Download Festival hygiene data. Load this file and run the syntax, or open a syntax window (see section 3.7) and type the following (remembering all of the full stops – the explanations of the code are surrounded by *s and don’t need to be typed).
DESCRIPTIVES
VARIABLES= day2/SAVE.
*This uses SPSS’s descriptives function on the variable day2 (instead of using the dialog box) to save the z-scores in the data editor (these will be saved as a variable called zday2).*
COMPUTE outlier1=abs(zday2).
EXECUTE.
*This creates a new variable in the data editor called outlier1, which contains the absolute values of the z-scores that we just created.*
RECODE
outlier1 (3.29 thru Highest=4) (2.58 thru Highest=3) (1.96 thru Highest=2) (Lowest thru 2=1).
EXECUTE.
*This recodes the variable called outlier1 according to the benchmarks I’ve described. So, if a value is greater than 3.29 it’s assigned a code of 4, if it’s between 2.58 and 3.29 then it’s assigned a code of 3, if it’s between 1.96 and 2.58 it’s assigned a code of 2, and if it’s less than 1.96 it gets a code of 1.*
VALUE LABELS outlier1
1 ‘Absolute z-score less than 2’ 2 ‘Absolute z-score greater than 1.96’ 3 ‘Absolute z-score greater than 2.58’ 4 ‘Absolute z-score greater than 3.29’.
*This assigns appropriate labels to the codes we defined above.*
FREQUENCIES
VARIABLES=outlier1
/ORDER=ANALYSIS.
*Finally, this syntax uses the frequencies facility of SPSS to produce a table telling us the percentage of 1s, 2s, 3s and 4s found in the variable outlier1.*
The table produced by this syntax is shown below. Look at the column labelled ‘Valid Percent’. We would expect to see 95% of cases with absolute value less than 1.96, 5% (or less) with an absolute value greater than 1.96, and 1% (or less) with an absolute value greater than 2.58. Finally, we’d expect no cases above 3.29 (well, these cases are significant outliers). For hygiene scores on day 2 of the festival, 93.2% of values had z-scores less than 1.96; put another way, 6.8% were above (looking at the table we get this figure by adding 4.5% + 1.5% + 0.8%). This is slightly more than the 5% we would expect in a normal distribution. Looking at values above 2.58, we would expect to find only 1%, but again here we have a higher value of 2.3% (1.5% + 0.8%). Finally, we find that 0.8% of cases were above 3.29 (so, 0.8% are significant outliers). This suggests that there may be too many outliers in this data set and we might want to do something about them!
Chapter 5: Exploring assumptions
5.4.1. Oh no, it’s that pesky frequency distribution again: checking normality visually
There is another useful graph that we can inspect to see if a distribution is normal called a P–P plot (probability–probability plot). This graph plots the cumulative probability of a variable against the cumulative probability of a particular distribution (in this case we would specify a normal distribution). What this means is that the data are ranked and sorted. Then for each rank the corresponding z-score is calculated. This is the expected value that the score should have in a normal distribution. Next the score itself is converted to a z-score (see section 1.7.4). The actual z-score is plotted against the expected z-score. If the data are normally distributed then the actual z-score will be the same as the expected z-score and you’ll get a lovely straight diagonal line. This ideal scenario is helpfully plotted on the graph and your job is to compare the data points to this line. If values fall on the diagonal of the plot then the variable is normally distributed, but deviations from the diagonal show deviations from normality.
To get a P–P plot use ANALYZE, DESCRIPTIVE STATISTICS, P-P PLOTS… to access the dialog box in Figure 5.2. There’s not a lot to say about this dialog box really because the default options will compare any variables selected to a normal distribution, which is what we want (although note that there is a drop-down list of different distributions against which you could compare your data). Select the three hygiene score variables in the variable list (click on the day 1 variable, then hold down Shift and select the day 3 variable and the day 2 scores will be selected as well). Transfer the selected variables to the box labelled Variables by clicking on ARROW. Click on OK to draw the graphs.
Figure 5.3 shows the histograms (from the self-test task) and the corresponding P–P plots. The first thing to note is that the data from day 1 look a lot more healthy since we’ve removed the data point that was mis-typed back in section 4.5. In fact the distribution is amazingly normal looking: it is nicely symmetrical and doesn’t seem too pointy or flat – these are good things! This is echoed by the P–P plot: note that the data points all fall very close to the ‘ideal’ diagonal line.
However, the distributions for days 2 and 3 are not nearly as symmetrical. In fact, they both look positively skewed. Again, this can be seen in the P–P plots by the data values deviating away from the diagonal. In general, what this seems to suggest is that by days 2 and 3, hygiene scores were much more clustered around the low end of the scale. Remember that the lower the score, the less hygienic the person is, so this suggests that generally people became smellier as the festival progressed. The skew occurs because a substantial minority insisted on upholding their levels of hygiene (against all odds!) over the course of the festival (baby wet-wipes are indispensable I find). However, these skewed distributions might cause us a problem if we want to use parametric tests. In the next section we’ll look at ways to try to quantify the skewness and kurtosis of these distributions.
5.4.2. Quantifying normality with numbers
It is all very well to look at histograms, but they are subjective and open to abuse (I can imagine researchers sitting looking at a completely distorted distribution and saying ‘yep, well Bob, that looks normal to me’, and Bob replying ‘yep, sure does’). Therefore, having inspected the distribution of hygiene scores visually, we can move on to look at ways to quantify the shape of the distributions and to look for outliers. To further explore the distribution of the variables, we can use the frequencies command (ANALYZE, DESCRIPTIVE STATISTICS, FREQUENCIES…). The main dialog box is shown in Figure 5.4. The variables in the data editor are listed on the left-hand side, and they can be transferred to the box labelled Variable(s) by clicking on a variable (or highlighting several with the mouse) and then clicking on ARROW. If a variable listed in the Variable(s) box is selected using the mouse, it can be transferred back to the variable list by clicking on the arrow button (which should now be pointing in the opposite direction). By default, SPSS produces a frequency distribution of all scores in table form. However, there are two other dialog boxes that can be selected that provide other options. The statistics dialog box is accessed by clicking on STATISTICS…, and the charts dialog box is accessed by clicking on CHARTS.
The statistics dialog box allows you to select several ways in which a distribution of scores can be described, such as measures of central tendency (mean, mode, median), measures of variability (range, standard deviation, variance, quartile splits), measures of shape (kurtosis and skewness). To describe the characteristics of the data we should select the mean, mode, median, standard deviation, variance and range. To check that a distribution of scores is normal, we need to look at the values of kurtosis and skewness (see section 1.7.1). The charts option provides a simple way to plot the frequency distribution of scores (as a bar chart, a pie chart or a histogram). We’ve already plotted histograms of our data so we don’t need to select these options, but you could use these options in future analyses. When you have selected the appropriate options, return to the main dialog box by clicking on CONTINUE. Once in the main dialog box, click on OK to run the analysis.
SPSS Output 5.1 shows the table of descriptive statistics for the three variables in this example. From this table, we can see that, on average, hygiene scores were 1.77 (out of 5) on day 1 of the festival, but went down to 0.96 and 0.98 on days 2 and 3 respectively. The other important measures for our purposes are the skewness and the kurtosis (see section 1.7.1), both of which have an associated standard error. The values of skewness and kurtosis should be zero in a normal distribution. Positive values of skewness indicate a pile-up of scores on the left of the distribution, whereas negative values indicate a pile-up on the right. Positive values of kurtosis indicate a pointy and heavy-tailed distribution, whereas negative values indicate a flat and light-tailed distribution. The further the value is from zero, the more likely it is that the data are not normally distributed. For day 1 the skew value is very close to zero (which is good) and kurtosis is a little negative. For days 2 and 3, though, there is a skewness of around 1 (positive skew).
Although the values of skew and kurtosis are informative, we can convert these values to z-scores. We saw in section 1.7.4 that a z-score is simply a score from a distribution that has a mean of 0 and a standard deviation of 1. We also saw that this distribution has known properties that we can use. Converting scores to a z-score is useful then because (1) we can compare skew and kurtosis values in different samples that used different measures, and (2) we can see how likely our values of skew and kurtosis are to occur. To transform any score to a z-score you simply subtract the mean of the distribution (in this case zero) and then divide by the standard deviation of the distribution (in this case we use the standard error). Skewness and kurtosis are converted to z-scores in exactly this way.
Zskewness = (S − 0) / SEskewness
Zkurtosis = (K − 0) / SEkurtosis
In the above equations, the values of S (skewness) and K (kurtosis) and their respective standard errors are produced by SPSS. These z-scores can be compared against values that you would expect to get by chance alone (i.e. known values for the normal distribution shown in the Appendix). So, an absolute value greater than 1.96 is significant at p < .05, above 2.58 is significant at p < .01 and absolute values above about 3.29 are significant at p < .001. Large samples will give rise to small standard errors and so when sample sizes are big, significant values arise from even small deviations from normality. In smallish samples it’s OK to look for values above 1.96; however, in large samples this criterion should be increased to the 2.58 one and in very large samples, because of the problem of small standard errors that I’ve described, no criterion should be applied! If you have a large sample (200 or more) it is more important to look at the shape of the distribution visually and to look at the value of the skewness and kurtosis statistics rather than calculate their significance.
For the hygiene scores, the z-score of skewness is -0.004/0.086 = 0.047 on day 1, 1.095/0.150 = 7.300 on day 2 and 1.033/0.218 = 4.739 on day 3. It is pretty clear then that although on day 1 scores are not at all skewed, on days 2 and 3 there is a very significant positive skew (as was evident from the histogram) – however, bear in mind what I just said about large samples! The kurtosis z-scores are: -0.410/0.172 = -2.38 on day 1, 0.822/0.299 = 2.75 on day 2 and 0.732/0.433 = 1.69 on day 3. These values indicate significant kurtosis (at p < .05) for all three days; however, because of the large sample, this isn’t surprising and so we can take comfort in the fact that all values of kurtosis are below our upper threshold of 3.29.
CRAMMING SAM’s Tips Skewness and kurtosis
• To check that the distribution of scores is approximately normal, we need to look at the values of skewness and kurtosis in the SPSS output.
• Positive values of skewness indicate too many low scores in the distribution, whereas negative values indicate a build-up of high scores.
• Positive values of kurtosis indicate a pointy and heavy-tailed distribution, whereas negative values indicate a flat and light-tailed distribution.
• The further the value is from zero, the more likely it is that the data are not normally distributed.
• You can convert these scores to z-scores by dividing by their standard error. If the resulting score (when you ignore the minus sign) is greater than 1.96 then it is significant (p < .05).
• Significance tests of skew and kurtosis should not be used in large samples (because they are likely to be significant even when skew and kurtosis are not too different from normal).
5.5. Testing whether a distribution is normal
Another way of looking at the problem is to see whether the distribution as a whole deviates from a comparable normal distribution. The Kolmogorov–Smirnov test and Shapiro–Wilk test do just this: they compare the scores in the sample to a normally distributed set of scores with the same mean and standard deviation. If the test is non-significant (p > .05) it tells us that the distribution of the sample is not significantly different from a normal distribution (i.e. it is probably normal). If, however, the test is significant (p < .05) then the distribution in question is significantly different from a normal distribution (i.e. it is non-normal). These tests seem great: in one easy procedure they tell us whether our scores are normally distributed (nice!). However, they have their limitations because with large sample sizes it is very easy to get significant results from small deviations from normality, and so a significant test doesn’t necessarily tell us whether the deviation from normality is enough to bias any statistical procedures that we apply to the data. I guess the take-home message is: by all means use these tests, but plot your data as well and try to make an informed decision about the extent of non-normality.
5.5.1. Doing the Kolmogorov–Smirnov test on SPSS
The Kolmogorov–Smirnov (K–S from now on; Figure 5.8) test can be accessed through the explore command (ANALYZE, DESCRIPTIVE STATISTICS, EXPLORE…). Figure 5.9 shows the dialog boxes for the explore command. First, enter any variables of interest in the box labelled Dependent List by highlighting them on the left-hand side and transferring them by clicking on ARROW. For this example, just select the exam scores and numeracy scores. It is also possible to select a factor (or grouping variable) by which to split the output (so, if you select Uni and transfer it to the box labelled Factor List, SPSS will produce exploratory analysis for each group – a bit like the split file command). If you click on STATISTICS… a dialog box appears, but the default option is fine (it will produce means, standard deviations and so on). The more interesting option for our current purposes is accessed by clicking on PLOTS… . In this dialog box select the option NORMALITY PLOTS WITH TESTS, and this will produce both the K–S test and some graphs called normal Q–Q plots. A Q–Q plot is very similar to the P-P plot that we encountered in section 5.4.1 except that it plots the quantiles of the data set instead of every individual score in the data. Quantiles are just values that split a data set into equal portions. We have already used quantiles without knowing it because quartiles (as in the interquartile range in section 1.7.3) are a special case of quantiles that split the data into four equal parts. However, you can have other quantiles such as percentiles (points that split the data into 100 equal parts), noniles (points that split the data into nine equal parts) and so on. In short, then, the Q–Q plot can be interpreted in the same way as a P–P plot but it will have less points on it because rather than plotting every single data point it plots only values that divide the data into equal parts (so, they can be easier to interpret if you have a lot of scores). By default, SPSS will produce boxplots (split according to group if a factor has been specified) and stem and leaf diagrams as well. Click on CONTINUE to return to the main dialog box and then click on OK to run the analysis.
5.5.2. Output from the explore procedure
The first table produced by SPSS contains descriptive statistics (mean etc.) and should have the same values as the tables obtained using the frequencies procedure. The important table is that of the K–S test (SPSS Output 5.4). This table includes the test statistic itself, the degrees of freedom (which should equal the sample size) and the significance value of this test. Remember that a significant value (Sig. less than .05) indicates a deviation from normality. For both numeracy and SPSS exam scores, the K–S test is highly significant, The first table produced by SPSS contains descriptive statistics (mean etc.) and should have the same values as the tables obtained using the frequencies procedure. The important table is that of the K–S test (SPSS Output 5.4). This table includes the test statistic itself, the degrees of freedom (which should equal the sample size) and the significance value of this test. Remember that a significant value (Sig. less than .05) indicates a deviation from normality. For both numeracy and SPSS exam scores, the K–S test is highly significant, indicating that both distributions are not normal. This result is likely to reflect the bimodal distribution found for exam scores, and the positively skewed distribution observed in the numeracy scores. However, these tests confirm that these deviations were significant. (But bear in mind that the sample is fairly big.)
As a final point, bear in mind that when we looked at the exam scores for separate groups, the distributions seemed quite normal; now if we’d asked for separate tests for the two universities (by placing Uni in the box labelled Factor List as in Figure 5.9) the K–S test might not have been significant. In fact if you try this out, you’ll get the table in SPSS Output 5.5, which shows that the percentages on the SPSS exam are indeed normal within the two groups (the values in the Sig. column are greater than .05). This is important because if our analysis involves comparing groups, then what’s important is not the overall distribution but the distribution in each group.
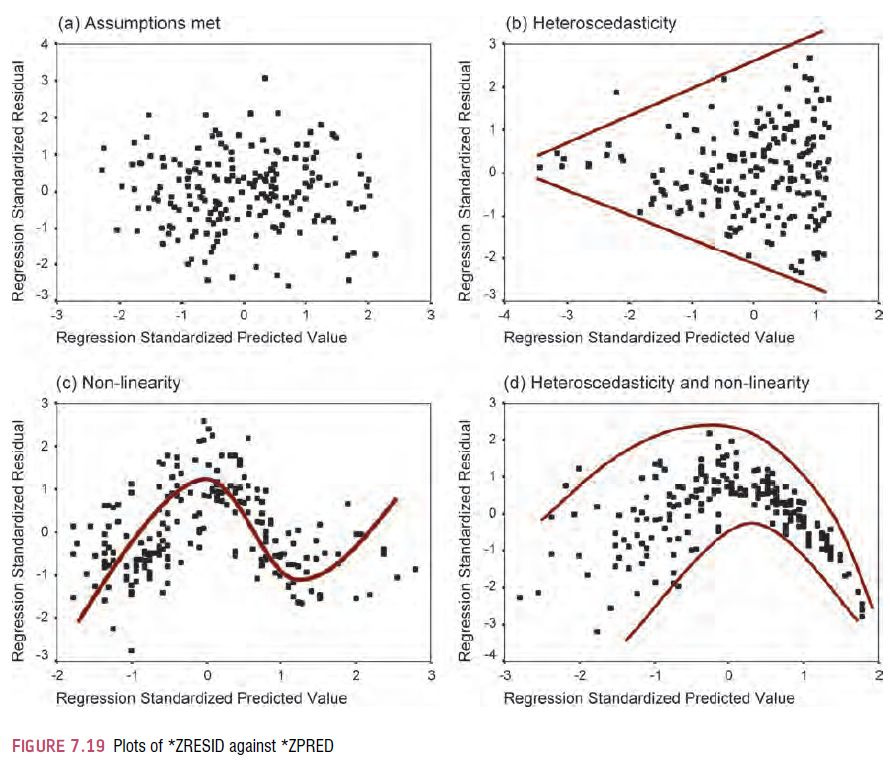
SPSS also produces a normal Q–Q plot for any variables specified (see Figure 5.10). The normal Q–Q chart plots the values you would expect to get if the distribution were normal (expected values) against the values actually seen in the data set (observed values). The expected values are a straight diagonal line, whereas the observed values are plotted as individual points. If the data are normally distributed, then the observed values (the dots on the chart) should fall exactly along the straight line (meaning that the observed values are the same as you would expect to get from a normally distributed data set). Any deviation of the dots from the line represents a deviation from normality. So, if the Q–Q plot looks like a straight line with a wiggly snake wrapped around it then you have some deviation from normality! Specifically, when the line sags consistently below the diagonal, or consistently rises above it, then this shows that the kurtosis differs from a normal distribution, and when the curve is S-shaped, the problem is skewness.
In both of the variables analysed we already know that the data are not normal, and these plots confirm this observation because the dots deviate substantially from the line. It is noteworthy that the deviation is greater for the numeracy scores, and this is consistent with the higher significance value of this variable on the K–S test.
5.6. Testing for homogeneity of variance
So far I’ve concentrated on the assumption of normally distributed data; however, at the beginning of this chapter I mentioned another assumption: homogeneity of variance. This assumption means that as you go through levels of one variable, the variance of the other should not change. If you’ve collected groups of data then this means that the variance of your outcome variable or variables should be the same in each of these groups. If you’ve collected continuous data (such as in correlational designs), this assumption means that the variance of one variable should be stable at all levels of the other variable. Let’s illustrate this with an example. An audiologist was interested in the effects of loud concerts on people’s hearing. So, she decided to send 10 people on tour with the loudest band she could find, Motörhead. These people went to concerts in Brixton (London), Brighton, Bristol, Edinburgh, Newcastle, Cardiff and Dublin and after each concert the audiologist measured the number of hours after the concert that these people had ringing in their ears.
Figure 5.11 shows the number of hours that each person had ringing in their ears after each concert (each person is represented by a circle). The horizontal lines represent the average number of hours that there was ringing in the ears after each concert and these means are connected by a line so that we can see the general trend of the data. Remember that for each concert, the circles are the scores from which the mean is calculated. Now, we can see in both graphs that the means increase as the people go to more concerts. So, after the first concert their ears ring for about 12 hours, but after the second they ring for about 15–20 hours, and by the final night of the tour, they ring for about 45–50 hours (2 days). So, there is a cumulative effect of the concerts on ringing in the ears. This pattern is found in both graphs; the difference between the graphs is not in terms of the means (which are roughly the same), but in terms of the spread of scores around the mean. If you look at the left-hand graph the spread of scores around the mean stays the same after each concert (the scores are fairly tightly packed around the mean). Put another way, if you measured the vertical distance between the lowest score and the highest score after the Brixton concert, and then did the same after the other concerts, all of these distances would be fairly similar. Although the means increase, the spread of scores for hearing loss is the same at each level of the concert variable (the spread of scores is the same after Brixton, Brighton, Bristol, Edinburgh, Newcastle, Cardiff and Dublin). This is what we mean by homogeneity of variance. The right-hand graph shows a different picture: if you look at the spread of scores after the Brixton concert, they are quite tightly packed around the mean (the vertical distance from the lowest score to the highest score is small), but after the Dublin show (for example) the scores are very spread out around the mean (the vertical distance from the lowest score to the highest score is large). This is an example of heterogeneity of variance: that is, at some levels of the concert variable the variance of scores is different to other levels (graphically, the vertical distance from the lowest to highest score is different after different concerts).
5.6.1. Levene’s test
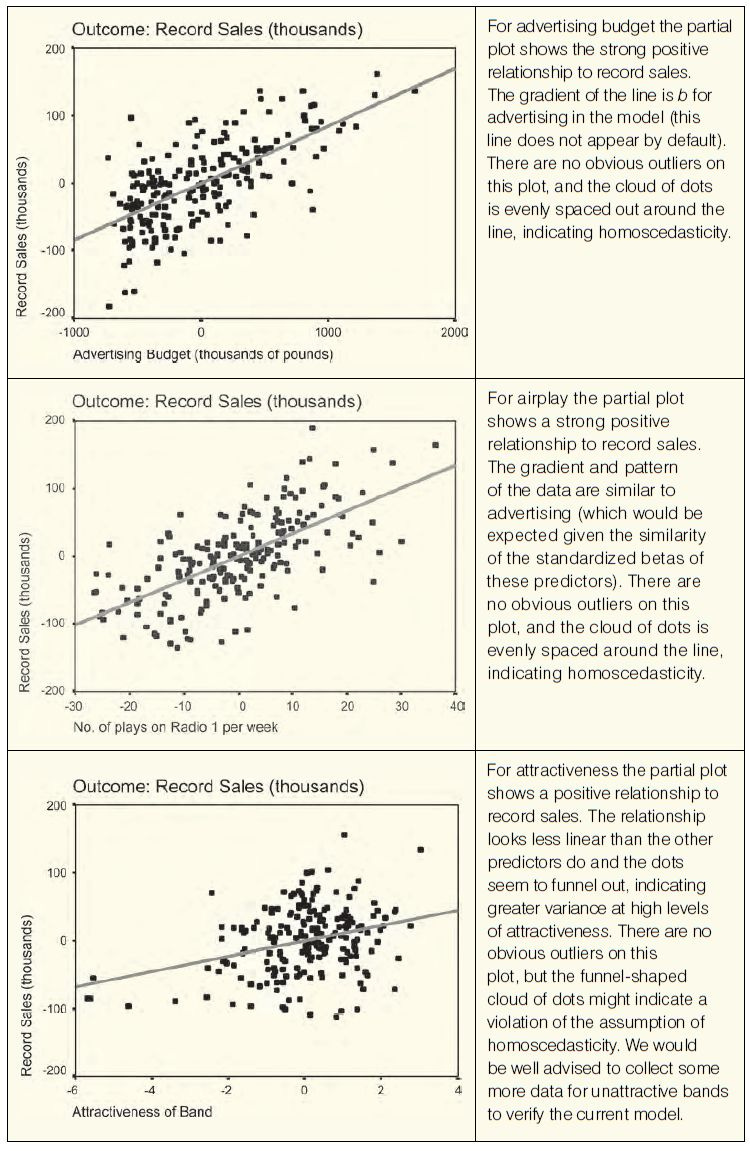
Hopefully you’ve got a grip of what homogeneity of variance actually means. Now, how do we test for it? Well, we could just look at the values of the variances and see whether they are similar. However, this approach would be very subjective and probably prone to academics thinking ‘Ooh look, the variance in one group is only 3000 times larger than the variance in the other: that’s roughly equal’. Instead, in correlational analysis such as regression we tend to use graphs (see section 7.8.7) and for groups of data we tend to use a test called Levene’s test (Levene, 1960). Levene’s test tests the null hypothesis that the variances in different groups are equal (i.e. the difference between the variances is zero). [...]
As with the K–S test (and other tests of normality), when the sample size is large, small differences in group variances can produce a Levene’s test that is significant (because, as we saw in Chapter 1, the power of the test is improved). A useful double check, therefore, is to look at Hartley’s FMax, also known as the variance ratio (Pearson & Hartley, 1954). This is the ratio of the variances between the group with the biggest variance and the group with the smallest variance.
JANE SUPERBRAIN 5.1 To transform or not to transform, that is the question
Not everyone agrees that transforming data is a good idea; for example, Glass, Peckham, and Sanders (1972) in a very extensive review commented that ‘the payoff of normalizing transformations in terms of more valid probability statements is low, and they are seldom considered to be worth the effort’ (p. 241). In which case, should we bother?
The issue is quite complicated (especially for this early in the book), but essentially we need to know whether the statistical models we apply perform better on transformed data than they do when applied to data that violate the assumption that the transformation corrects. If a statistical model is still accurate even when its assumptions are broken it is said to be a robust test (section 5.7.4). I’m not going to discuss whether particular tests are robust here, but I will discuss the issue for particular tests in their respective chapters. The question of whether to transform is linked to this issue of robustness (which in turn is linked to what test you are performing on your data).
A good case in point is the F-test in ANOVA (see Chapter 10), which is often claimed to be robust (Glass et al., 1972). Early findings suggested that F performed as it should in skewed distributions and that transforming the data helped as often as it hindered the accuracy of F (Games & Lucas, 1966). However, in a lively but informative exchange Levine and Dunlap (1982) showed that transformations of skew did improve the performance of F; however, in a response Games (1983) argued that their conclusion was incorrect, which Levine and Dunlap (1983) contested in a response to the response. Finally, in a response to the response of the response, Games (1984) pointed out several important questions to consider:
1. The central limit theorem (section 2.5.1) tells us that in big samples the sampling distribution will be normal regardless, and this is what’s actually important so the debate is academic in anything other than small samples. Lots of early research did indeed show that with samples of 40 the normality of the sampling distribution was, as predicted, normal. However, this research focused on distributions with light tails and subsequent work has shown that with heavy-tailed distributions larger samples would be necessary to invoke the central limit theorem (Wilcox, 2005). This research suggests that transformations might be useful for such distributions.
2. By transforming the data you change the hypothesis being tested (when using a log transformation and comparing means you change from comparing arithmetic means to comparing geometric means). Transformation also means that you’re now addressing a different construct to the one originally measured, and this has obvious implications for interpreting that data (Grayson, 2004).
3. In small samples it is tricky to determine normality one way or another (tests such as K–S will have low power to detect deviations from normality and graphs will be hard to interpret with so few data points).
4. The consequences for the statistical model of applying the ‘wrong’ transformation could be worse than the consequences of analysing the untransformed scores.
As we will see later in the book, there is an extensive library of robust tests that can be used and which have considerable benefits over transforming data. The definitive guide to these is Wilcox’s (2005) outstanding book.
5.7.4. When it all goes horribly wrong
It’s very easy to think that transformations are the answers to all of your broken assumption prayers. However, as we have seen, there are reasons to think that transformations are not necessarily a good idea (see Jane Superbrain Box 5.1) and even if you think that they are they do not always solve the problem, and even when they do solve the problem they often create different problems in the process. This happens more frequently than you might imagine (messy data are the norm).
If you find yourself in the unenviable position of having irksome data then there are some other options available to you (other than sticking a big samurai sword through your head). The first is to use a test that does not rely on the assumption of normally distributed data and as you go through the various chapters of this book I’ll point out these tests – there is also a whole chapter dedicated to them later on. [7] One thing that you will quickly discover about non-parametric tests is that they have been developed for only a fairly limited range of situations. So, happy days if you want to compare two means, but sad lonely days listening to Joy Division if you have a complex experimental design.
A much more promising approach is to use robust methods (which I mentioned in Jane Superbrain Box 5.1). These tests have developed as computers have got more sophisticated (doing these tests without computers would be only marginally less painful than ripping off your skin and diving into a bath of salt). How these tests work is beyond the scope of this book (and my brain) but two simple concepts will give you the general idea. Some of these procedures use a trimmed mean. A trimmed mean is simply a mean based on the distribution of scores after some percentage of scores has been removed from each extreme of the distribution. So, a 10% trimmed mean will remove 10% of scores from the top and bottom before the mean is calculated. We saw in Chapter 2 that the accuracy of the mean depends on a symmetrical distribution, but a trimmed mean produces accurate results even when the distribution is not symmetrical, because by trimming the ends of the distribution we remove outliers and skew that bias the mean. Some robust methods work by taking advantage of the properties of the trimmed mean.
The second general procedure is the bootstrap (Efron & Tibshirani, 1993). The idea of the bootstrap is really very simple and elegant. The problem that we have is that we don’t know the shape of the sampling distribution, but normality in our data allows us to infer that the sampling distribution is normal (and hence we can know the probability of a particular test statistic occurring). Lack of normality prevents us from knowing the shape of the sampling distribution unless we have big samples (but see Jane Superbrain Box 5.1). Bootstrapping gets around this problem by estimating the properties of the sampling distribution from the sample data. In effect, the sample data are treated as a population from which smaller samples (called bootstrap samples) are taken (putting the data back before a new sample is drawn). The statistic of interest (e.g. the mean) is calculated in each sample, and by taking many samples the sampling distribution can be estimated (rather like in Figure 2.7). The standard error of the statistic is estimated from the standard deviation of this sampling distribution created from the bootstrap samples. From this standard error, confidence intervals and significance tests can be computed. This is a very neat way of getting around the problem of not knowing the shape of the sampling distribution.
These techniques sound pretty good don’t they? It might seem a little strange then that I haven’t written a chapter on them. The reason why is that SPSS does not do most of them, which is something that I hope it will correct sooner rather than later. However, thanks to Rand Wilcox you can do them using a free statistics program called R (www.r-project.org) and a non-free program called S-Plus. Wilcox provides a very comprehensive review of robust methods in his excellent book Introduction to robust estimation and hypothesis testing (2005) and has written programs to run these methods using R. Among many other things, he has files to run robust versions of many tests discussed in this book: ANOVA, ANCOVA, correlation and multiple regression. If there is a robust method, it is likely to be in his book, and he will have written a macro procedure to run it! You can also download these macros from his website. [8] There are several good introductory books to tell you how to use R also (e.g. Dalgaard, 2002). There is also an SPSS plugin that you can download (http://www.spss.com/devcentral/index.cfm?pg=plugins) that allows you to use R commands from the SPSS syntax window. With this plugin you can analyse an open SPSS data file using the robust methods of R. These analyses are quite technical so I don’t discuss them in the book, but if you’d like to know more see Oliver Twisted.
[7] For convenience a lot of textbooks refer to these tests as non-parametric tests or assumption-free tests and stick them in a separate chapter. Actually neither of these terms are particularly accurate (e.g. none of these tests is assumption-free) but in keeping with tradition I’ve put them in a chapter (15) on their own ostracized from their ‘parametric’ counterparts and feeling lonely.
Chapter 6: Correlation
6.3.2. Standardization and the correlation coefficient
To overcome the problem of dependence on the measurement scale, we need to convert the covariance into a standard set of units. This process is known as standardization. A very basic form of standardization would be to insist that all experiments use the same units of measurement, say metres – that way, all results could be easily compared. However, what happens if you want to measure attitudes – you’d be hard pushed to measure them in metres! Therefore, we need a unit of measurement into which any scale of measurement can be converted. The unit of measurement we use is the standard deviation. We came across this measure in section 2.4.1 and saw that, like the variance, it is a measure of the average deviation from the mean. If we divide any distance from the mean by the standard deviation, it gives us that distance in standard deviation units. For example, for the data in Table 6.1, the standard deviation for the number of packets bought is approximately 3.0 (the exact value is 2.91). In Figure 6.2 we can see that the observed value for participant 1 was 3 packets less than the mean (so, there was an error of -3 packets of sweets). If we divide this deviation, -3, by the standard deviation, which is approximately 3, then we get a value of -1. This tells us that the difference between participant 1’s score and the mean was -1 standard deviation. So, we can express the deviation from the mean for a participant in standard units by dividing the observed deviation by the standard deviation.
It follows from this logic that if we want to express the covariance in a standard unit of measurement we can simply divide by the standard deviation. However, there are two variables and, hence, two standard deviations. Now, when we calculate the covariance we actually calculate two deviations (one for each variable) and then multiply them. Therefore, we do the same for the standard deviations: we multiply them and divide by the product of this multiplication. The standardized covariance is known as a correlation coefficient and is defined by equation (6.3) in which sx is the standard deviation of the first variable and sy is the standard deviation of the second variable (all other letters are the same as in the equation defining covariance):
r = covxy/sxsy = Σ(xi – x̄) (yi – ȳ) / (N – 1)sxsy (6.3)
The coefficient in equation (6.3) is known as the Pearson product-moment correlation coefficient or Pearson correlation coefficient (for a really nice explanation of why it was originally called the ‘product-moment’ correlation see Miles & Banyard, 2007) and was invented by Karl Pearson (see Jane Superbrain Box 6.1). [2] If we look back at Table 6.1 we see that the standard deviation for the number of adverts watched (sx) was 1.67, and for the number of packets of crisps bought (sy) was 2.92. If we multiply these together we get 1.67 × 2.92 = 4.88. Now, all we need to do is take the covariance, which we calculated a few pages ago as being 4.25, and divide by these multiplied standard deviations. This gives us r = 4.25/4.88 = .87.
By standardizing the covariance we end up with a value that has to lie between -1 and +1 (if you find a correlation coefficient less than -1 or more than +1 you can be sure that something has gone hideously wrong!). A coefficient of +1 indicates that the two variables are perfectly positively correlated, so as one variable increases, the other increases by a proportionate amount. Conversely, a coefficient of -1 indicates a perfect negative relationship: if one variable increases, the other decreases by a proportionate amount. A coefficient of zero indicates no linear relationship at all and so if one variable changes, the other stays the same. We also saw in section 2.6.4 that because the correlation coefficient is a standardized measure of an observed effect, it is a commonly used measure of the size of an effect and that values of ±.1 represent a small effect, ±.3 is a medium effect and ±.5 is a large effect (although I re-emphasize my caveat that these canned effect sizes are no substitute for interpreting the effect size within the context of the research literature).
6.5.2. Pearson’s correlation coefficient
6.5.2.1. A ssumptions of Pearson’s r
Pearson’s correlation coefficient was described in full at the beginning of this chapter. Pearson’s correlation requires only that data are interval (see section 1.5.1.2) for it to be an accurate measure of the linear relationship between two variables. However, if you want to establish whether the correlation coefficient is significant, then more assumptions are required: for the test statistic to be valid the sampling distribution has to be normally distributed and as we saw in Chapter 5 we assume that it is if our sample data are normally distributed (or if we have a large sample). Although typically, to assume that the sampling distribution is normal, we would want both variables to be normally distributed, there is one exception to this rule: one of the variables can be a categorical variable provided there are only two categories (in fact, if you look at section 6.5.5 you’ll see that this is the same as doing a t-test, but I’m jumping the gun a bit). In any case, if your data are non-normal (see Chapter 5) or are not measured at the interval level then you should deselect the Pearson tick-box.
6.5.5. Biserial and point-biserial correlations
The biserial and point–biserial correlation coefficients are distinguished by only a conceptual difference yet their statistical calculation is quite different. These correlation coefficients are used when one of the two variables is dichotomous (i.e. it is categorical with only two categories). An example of a dichotomous variable is being pregnant, because a woman can be either pregnant or not (she cannot be ‘a bit pregnant’). Often it is necessary to investigate relationships between two variables when one of the variables is dichotomous. The difference between the use of biserial and point–biserial correlations depends on whether the dichotomous variable is discrete or continuous. This difference is very subtle. A discrete, or true, dichotomy is one for which there is no underlying continuum between the categories. An example of this is whether someone is dead or alive: a person can be only dead or alive, they can’t be ‘a bit dead’. Although you might describe a person as being ‘half-dead’ – especially after a heavy drinking session – they are clearly still alive if they are still breathing! Therefore, there is no continuum between the two categories. However, it is possible to have a dichotomy for which a continuum does exist. An example is passing or failing a statistics test: some people will only just fail while others will fail by a large margin; likewise some people will scrape a pass while others will clearly excel. So although participants fall into only two categories there is clearly an underlying continuum along which people lie. Hopefully, it is clear that in this case there is some kind of continuum underlying the dichotomy, because some people passed or failed more dramatically than others. The point–biserial correlation coefficient (rpb) is used when one variable is a discrete dichotomy (e.g. pregnancy), whereas the biserial correlation coefficient (rb) is used when one variable is a continuous dichotomy (e.g. passing or failing an exam). The biserial correlation coefficient cannot be calculated directly in SPSS: first you must calculate the point–biserial correlation coefficient and then use an equation to adjust that figure.
Imagine that I was interested in the relationship between the gender of a cat and how much time it spent away from home (what can I say? I love cats so these things interest me). I had heard that male cats disappeared for substantial amounts of time on long-distance roams around the neighbourhood (something about hormones driving them to find mates) whereas female cats tended to be more homebound. So, I used this as a purr-fect (sorry!) excuse to go and visit lots of my friends and their cats. I took a note of the gender of the cat and then asked the owners to note down the number of hours that their cat was absent from home over a week. Clearly the time spent away from home is measured at an interval level – and let’s assume it meets the other assumptions of parametric data – while the gender of the cat is discrete dichotomy. A point–biserial correlation has to be calculated and this is simply a Pearson correlation when the dichotomous variable is coded with 0 for one category and 1 for the other (actually you can use any values and SPSS will change the lower one to 0 and the higher one to 1 when it does the calculations). So, to conduct these correlations in SPSS assign the Gender variable a coding scheme as described in section 3.4.2.3 (in the saved data the coding is 1 for a male and 0 for a female). The time variable simply has time in hours recorded as normal. These data are in the file pbcorr.sav.
6.6. Partial correlation
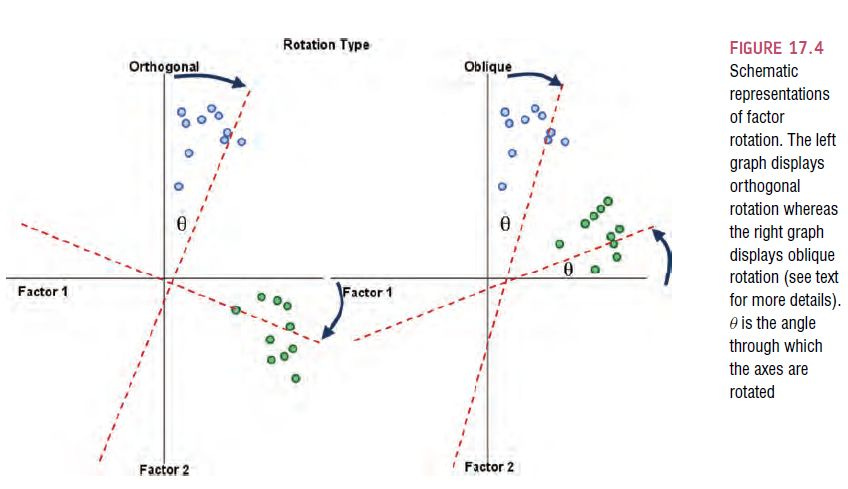
6.6.1. The theory behind part and partial correlation
I mentioned earlier that there is a type of correlation that can be done that allows you to look at the relationship between two variables when the effects of a third variable are held constant. For example, analyses of the exam anxiety data (in the file ExamAnxiety.sav) showed that exam performance was negatively related to exam anxiety, but positively related to revision time, and revision time itself was negatively related to exam anxiety. This scenario is complex, but given that we know that revision time is related to both exam anxiety and exam performance, then if we want a pure measure of the relationship between exam anxiety and exam performance we need to take account of the influence of revision time. Using the values of R² for these relationships, we know that exam anxiety accounts for 19.4% of the variance in exam performance, that revision time accounts for 15.7% of the variance in exam performance, and that revision time accounts for 50.2% of the variance in exam anxiety. If revision time accounts for half of the variance in exam anxiety, then it seems feasible that at least some of the 19.4% of variance in exam performance that is accounted for by anxiety is the same variance that is accounted for by revision time. As such, some of the variance in exam performance explained by exam anxiety is not unique and can be accounted for by revision time. A correlation between two variables in which the effects of other variables are held constant is known as a partial correlation.
Let’s return to our example of exam scores, revision time and exam anxiety to illustrate the principle behind partial correlation (Figure 6.9). In part 1 of the diagram there is a box for exam performance that represents the total variation in exam scores (this value would be the variance of exam performance). There is also a box that represents the variation in exam anxiety (again, this is the variance of that variable). We know already that exam anxiety and exam performance share 19.4% of their variation (this value is the correlation coefficient squared). Therefore, the variations of these two variables overlap (because they share variance) creating a third box (the one with diagonal lines). The overlap of the boxes representing exam performance and exam anxiety is the common variance. Likewise, in part 2 of the diagram the shared variation between exam performance and revision time is illustrated. Revision time shares 15.7% of the variation in exam scores. This shared variation is represented by the area of overlap (filled with diagonal lines). We know that revision time and exam anxiety also share 50% of their variation; therefore, it is very probable that some of the variation in exam performance shared by exam anxiety is the same as the variance shared by revision time.
Part 3 of the diagram shows the complete picture. The first thing to note is that the boxes representing exam anxiety and revision time have a large overlap (this is because they share 50% of their variation). More important, when we look at how revision time and anxiety contribute to exam performance we see that there is a portion of exam performance that is shared by both anxiety and revision time (the dotted area). However, there are still small chunks of the variance in exam performance that are unique to the other two variables. So, although in part 1 exam anxiety shared a large chunk of variation in exam performance, some of this overlap is also shared by revision time. If we remove the portion of variation that is also shared by revision time, we get a measure of the unique relationship between exam performance and exam anxiety. We use partial correlations to find out the size of the unique portion of variance. Therefore, we could conduct a partial correlation between exam anxiety and exam performance while ‘controlling’ for the effect of revision time. Likewise, we could carry out a partial correlation between revision time and exam performance while ‘controlling’ for the effects of exam anxiety.
6.6.3. Semi-partial (or part) correlations
In the next chapter, we will come across another form of correlation known as a semi-partial correlation (also referred to as a part correlation). While I’m babbling on about partial correlations it is worth my explaining the difference between this type of correlation and a semipartial correlation. When we do a partial correlation between two variables, we control for the effects of a third variable. Specifically, the effect that the third variable has on both variables in the correlation is controlled. In a semi-partial correlation we control for the effect that the third variable has on only one of the variables in the correlation. Figure 6.12 illustrates this principle for the exam performance data. The partial correlation that we calculated took account not only of the effect of revision on exam performance, but also of the effect of revision on anxiety. If we were to calculate the semi-partial correlation for the same data, then this would control for only the effect of revision on exam performance (the effect of revision on exam anxiety is ignored). Partial correlations are most useful for looking at the unique relationship between two variables when other variables are ruled out. Semi-partial correlations are, therefore, useful when trying to explain the variance in one particular variable (an outcome) from a set of predictor variables. (Bear this in mind when you read Chapter 7.)
Chapter 7: Regression
7.2.3. Assessing the goodness of fit: sums of squares, R and R²
Once Nephwick the Line Finder has found the line of best fit it is important that we assess how well this line fits the actual data (we assess the goodness of fit of the model). We do this because even though this line is the best one available, it can still be a lousy fit to the data! In section 2.4.2 we saw that one measure of the adequacy of a model is the sum of squared differences (or more generally we assess models using equation (7.3) below). If we want to assess the line of best fit, we need to compare it against something, and the thing we choose is the most basic model we can find. So we use equation (7.3) to calculate the fit of the most basic model, and then the fit of the best model (the line of best fit), and basically if the best model is any good then it should fit the data significantly better than our basic model:
deviation = Σ(observed − model)² (7.3)
This is all quite abstract so let’s look at an example. Imagine that I was interested in predicting record sales (Y) from the amount of money spent advertising that record (X). One day my boss came in to my office and said ‘Andy, I know you wanted to be a rock star and you’ve ended up working as my stats-monkey, but how many records will we sell if we spend £100,000 on advertising?’ If I didn’t have an accurate model of the relationship between record sales and advertising, what would my best guess be? Well, probably the best answer I could give would be the mean number of record sales (say, 200,000) because on average that’s how many records we expect to sell. This response might well satisfy a brainless record company executive (who didn’t offer my band a record contract). However, what if he had asked ‘How many records will we sell if we spend £1 on advertising?’ Again, in the absence of any accurate information, my best guess would be to give the average number of sales (200,000). There is a problem: whatever amount of money is spent on advertising I always predict the same levels of sales. As such, the mean is a model of ‘no relationship’ at all between the variables. It should be pretty clear then that the mean is fairly useless as a model of a relationship between two variables – but it is the simplest model available.
So, as a basic strategy for predicting the outcome, we might choose to use the mean, because on average it will be a fairly good guess of an outcome. Using the mean as a model, we can calculate the difference between the observed values, and the values predicted by the mean (equation (7.3)). We saw in section 2.4.2 that we square all of these differences to give us the sum of squared differences. This sum of squared differences is known as the total sum of squares (denoted SST) because it is the total amount of differences present when the most basic model is applied to the data. This value represents how good the mean is as a model of the observed data. Now, if we fit the more sophisticated model to the data, such as a line of best fit, we can again work out the differences between this new model and the observed data (again using equation (7.3)). In the previous section we saw that the method of least squares finds the best possible line to describe a set of data by minimizing the difference between the model fitted to the data and the data themselves. However, even with this optimal model there is still some inaccuracy, which is represented by the differences between each observed data point and the value predicted by the regression line. As before, these differences are squared before they are added up so that the directions of the differences do not cancel out. The result is known as the sum of squared residuals or residual sum of squares (SSR). This value represents the degree of inaccuracy when the best model is fitted to the data. We can use these two values to calculate how much better the regression line (the line of best fit) is than just using the mean as a model (i.e. how much better is the best possible model than the worst model?). The improvement in prediction resulting from using the regression model rather than the mean is calculated by calculating the difference between SST and SSR. This difference shows us the reduction in the inaccuracy of the model resulting from fitting the regression model to the data. This improvement is the model sum of squares (SSM). Figure 7.4 shows each sum of squares graphically.
If the value of SSM is large then the regression model is very different from using the mean to predict the outcome variable. This implies that the regression model has made a big improvement to how well the outcome variable can be predicted. However, if SSM is small then using the regression model is little better than using the mean (i.e. the regression model is no better than taking our ‘best guess’). A useful measure arising from these sums of squares is the proportion of improvement due to the model. This is easily calculated by dividing the sum of squares for the model by the total sum of squares. The resulting value is called R² and to express this value as a percentage you should multiply it by 100. R² represents the amount of variance in the outcome explained by the model (SSM) relative to how much variation there was to explain in the first place (SST). Therefore, as a percentage, it represents the percentage of the variation in the outcome that can be explained by the model:
R² = SSM/SST (7.4)
This R² is the same as the one we met in Chapter 6 (section 6.5.2.3) and you might have noticed that it is interpreted in the same way. Therefore, in simple regression we can take the square root of this value to obtain Pearson’s correlation coefficient. As such, the correlation coefficient provides us with a good estimate of the overall fit of the regression model, and R² provides us with a good gauge of the substantive size of the relationship. A second use of the sums of squares in assessing the model is through the F-test. I mentioned way back in Chapter 2 that test statistics (like F) are usually the amount of systematic variance divided by the amount of unsystematic variance, or, put another way, the model compared against the error in the model. This is true here: F is based upon the ratio of the improvement due to the model (SSM) and the difference between the model and the observed data (SSR). Actually, because the sums of squares depend on the number of differences that we have added up, we use the average sums of squares (referred to as the mean squares or MS). To work out the mean sums of squares we divide by the degrees of freedom (this is comparable to calculating the variance from the sums of squares – see section 2.4.2). For SSM the degrees of freedom are simply the number of variables in the model, and for SSR they are the number of observations minus the number of parameters being estimated (i.e. the number of beta coefficients including the constant). The result is the mean squares for the model (MSM) and the residual mean squares (MSR). At this stage it isn’t essential that you understand how the mean squares are derived (it is explained in Chapter 10). However, it is important that you understand that the F-ratio (equation (7.5)) is a measure of how much the model has improved the prediction of the outcome compared to the level of inaccuracy of the model:
F = MSM/MSR (7.5)
If a model is good, then we expect the improvement in prediction due to the model to be large (so, MSM will be large) and the difference between the model and the observed data to be small (so, MSR will be small). In short, a good model should have a large F-ratio (greater than 1 at least) because the top of equation (7.5) will be bigger than the bottom. The exact magnitude of this F-ratio can be assessed using critical values for the corresponding degrees of freedom (as in the Appendix).
7.2.4. Assessing individual predictors
We’ve seen that the predictor in a regression model has a coefficient (b1), which in simple regression represents the gradient of the regression line. The value of b represents the change in the outcome resulting from a unit change in the predictor. If the model was useless at predicting the outcome, then if the value of the predictor changes, what might we expect the change in the outcome to be? Well, if the model is very bad then we would expect the change in the outcome to be zero. Think back to Figure 7.4 (see the panel representing SST) in which we saw that using the mean was a very bad way of predicting the outcome. In fact, the line representing the mean is flat, which means that as the predictor variable changes, the value of the outcome does not change (because for each level of the predictor variable, we predict that the outcome will equal the mean value). The important point here is that a bad model (such as the mean) will have regression coefficients of 0 for the predictors. A regression coefficient of 0 means: (1) a unit change in the predictor variable results in no change in the predicted value of the outcome (the predicted value of the outcome does not change at all); and (2) the gradient of the regression line is 0, meaning that the regression line is flat. Hopefully, you’ll see that it logically follows that if a variable significantly predicts an outcome, then it should have a b-value significantly different from zero. This hypothesis is tested using a t-test (see Chapter 9). The t-statistic tests the null hypothesis that the value of b is 0: therefore, if it is significant we gain confidence in the hypothesis that the b-value is significantly different from 0 and that the predictor variable contributes significantly to our ability to estimate values of the outcome.
Like F, the t-statistic is also based on the ratio of explained variance against unexplained variance or error. Well, actually, what we’re interested in here is not so much variance but whether the b we have is big compared to the amount of error in that estimate. To estimate how much error we could expect to find in b we use the standard error. The standard error tells us something about how different b-values would be across different samples. We could take lots and lots of samples of data regarding record sales and advertising budgets and calculate the b-values for each sample. We could plot a frequency distribution of these samples to discover whether the b-values from all samples would be relatively similar, or whether they would be very different (think back to section 2.5.1). We can use the standard deviation of this distribution (known as the standard error) as a measure of the similarity of b-values across samples. If the standard error is very small, then it means that most samples are likely to have a b-value similar to the one in our sample (because there is little variation across samples). The t-test tells us whether the b-value is different from 0 relative to the variation in b-values across samples. When the standard error is small even a small deviation from zero can reflect a meaningful difference because b is representative of the majority of possible samples.
Equation (7.6) shows how the t-test is calculated and you’ll find a general version of this equation in Chapter 9 (equation (9.1)). The bexpected is simply the value of b that we would expect to obtain if the null hypothesis were true. I mentioned earlier that the null hypothesis is that b is 0 and so this value can be replaced by 0. The equation simplifies to become the observed value of b divided by the standard error with which it is associated:
t = bobserved − bexpected/SEb (7.6)
= bobserved/SEb
The values of t have a special distribution that differs according to the degrees of freedom for the test. In regression, the degrees of freedom are N − p − 1, where N is the total sample size and p is the number of predictors. In simple regression when we have only one predictor, this reduces down to N − 2. Having established which t-distribution needs to be used, the observed value of t can then be compared to the values that we would expect to find if there was no effect (i.e. b = 0): if t is very large then it is unlikely to have occurred when there is no effect (these values can be found in the Appendix). SPSS provides the exact probability that the observed value or a larger value of t would occur if the value of b was, in fact, 0. As a general rule, if this observed significance is less than .05, then scientists assume that b is significantly different from 0; put another way, the predictor makes a significant contribution to predicting the outcome.
7.4. Interpreting a simple regression
7.4.1. Overall fit of the model
The first table provided by SPSS is a summary of the model (SPSS Output 7.1). This summary table provides the value of R and R² for the model that has been derived. For these data, R has a value of .578 and because there is only one predictor, this value represents the simple correlation between advertising and record sales (you can confirm this by running a correlation using what you were taught in Chapter 6). The value of R² is .335, which tells us that advertising expenditure can account for 33.5% of the variation in record sales. In other words, if we are trying to explain why some records sell more than others, we can look at the variation in sales of different records. There might be many factors that can explain this variation, but our model, which includes only advertising expenditure, can explain approximately 33% of it. This means that 66% of the variation in record sales cannot be explained by advertising alone. Therefore, there must be other variables that have an influence also.
The next part of the output (SPSS Output 7.2) reports an analysis of variance (ANOVA – see Chapter 10). The summary table shows the various sums of squares described in Figure 7.4 and the degrees of freedom associated with each. From these two values, the average sums of squares (the mean squares) can be calculated by dividing the sums of squares by the associated degrees of freedom. The most important part of the table is the F-ratio, which is calculated using equation (7.5), and the associated significance value of that F-ratio. For these data, F is 99.59, which is significant at p < .001 (because the value in the column labelled Sig. is less than .001). This result tells us that there is less than a 0.1% chance that an F-ratio this large would happen if the null hypothesis were true. Therefore, we can conclude that our regression model results in significantly better prediction of record sales than if we used the mean value of record sales. In short, the regression model overall predicts record sales significantly well.
7.5.2. Sums of squares, R and R²
When we have several predictors, the partitioning of sums of squares is the same as in the single variable case except that the model we refer to takes the form of equation (7.9) rather than simply being a 2-D straight line. Therefore, SST can be calculated that represents the difference between the observed values and the mean value of the outcome variable. SSR still represents the difference between the values of Y predicted by the model and the observed values. Finally, SSM can still be calculated and represents the difference between the values of Y predicted by the model and the mean value. Although the computation of these values is much more complex than in simple regression, conceptually these values are the same.
When there are several predictors it does not make sense to look at the simple correlation coefficient and instead SPSS produces a multiple correlation coefficient (labelled Multiple R). Multiple R is the correlation between the observed values of Y and the values of Y predicted by the multiple regression model. Therefore, large values of the multiple R represent a large correlation between the predicted and observed values of the outcome. A multiple R of 1 represents a situation in which the model perfectly predicts the observed data. As such, multiple R is a gauge of how well the model predicts the observed data. It follows that the resulting R² can be interpreted in the same way as simple regression: it is the amount of variation in the outcome variable that is accounted for by the model.
7.6.1.1. Outliers and residuals
An outlier is a case that differs substantially from the main trend of the data (see Jane Superbrain Box 4.1). Figure 7.8 shows an example of such a case in regression. Outliers can cause your model to be biased because they affect the values of the estimated regression coefficients. For example, Figure 7.8 uses the same data as Figure 7.3 except that the score of one participant has been changed to be an outlier (in this case a person who was very calm in the presence of a very big spider). The change in this one point has had a dramatic effect on the regression model chosen to fit the data. With the outlier present, the regression model changes: its gradient is reduced (the line becomes flatter) and the intercept increases (the new line will cross the Y-axis at a higher point). It should be clear from this diagram that it is important to try to detect outliers to see whether the model is biased in this way.
How do you think that you might detect an outlier? Well, we know that an outlier, by its nature, is very different from all of the other scores. This being true, do you think that the model will predict that person’s score very accurately? The answer is no: looking at Figure 7.8 it is evident that even though the outlier has biased the model, the model still predicts that one value very badly (the regression line is long way from the outlier). Therefore, if we were to work out the differences between the data values that were collected, and the values predicted by the model, we could detect an outlier by looking for large differences. This process is the same as looking for cases that the model predicts inaccurately. The differences between the values of the outcome predicted by the model and the values of the outcome observed in the sample are known as residuals. These residuals represent the error present in the model. If a model fits the sample data well then all residuals will be small (if the model was a perfect fit of the sample data – all data points fall on the regression line – then all residuals would be zero). If a model is a poor fit of the sample data then the residuals will be large. Also, if any cases stand out as having a large residual, then they could be outliers.
The normal or unstandardized residuals described above are measured in the same units as the outcome variable and so are difficult to interpret across different models. What we can do is to look for residuals that stand out as being particularly large. However, we cannot define a universal cut-off point for what constitutes a large residual. To overcome this problem, we use standardized residuals, which are the residuals divided by an estimate of their standard deviation. We came across standardization in section 6.3.2 as a means of converting variables into a standard unit of measurement (the standard deviation); we also came across z-scores (see section 1.7.4) in which variables are converted into standard deviation units (i.e. they’re converted into scores that are distributed around a mean of 0 with a standard deviation of 1). By converting residuals into z-scores (standardized residuals) we can compare residuals from different models and use what we know about the properties of z-scores to devise universal guidelines for what constitutes an acceptable (or unacceptable) value. For example, we know from Chapter 1 that in a normally distributed sample, 95% of z-scores should lie between -1.96 and +1.96, 99% should lie between -2.58 and +2.58, and 99.9% (i.e. nearly all of them) should lie between -3.29 and +3.29. Some general rules for standardized residuals are derived from these facts: (1) standardized residuals with an absolute value greater than 3.29 (we can use 3 as an approximation) are cause for concern because in an average sample case a value this high is unlikely to happen by chance; (2) if more than 1% of our sample cases have standardized residuals with an absolute value greater than 2.58 (we usually just say 2.5) there is evidence that the level of error within our model is unacceptable (the model is a fairly poor fit of the sample data); and (3) if more than 5% of cases have standardized residuals with an absolute value greater than 1.96 (we can use 2 for convenience) then there is also evidence that the model is a poor representation of the actual data.
A third form of residual is the Studentized residual, which is the unstandardized residual divided by an estimate of its standard deviation that varies point by point. These residuals have the same properties as the standardized residuals but usually provide a more precise estimate of the error variance of a specific case.
7.6.1.2. Influential cases
As well as testing for outliers by looking at the error in the model, it is also possible to look at whether certain cases exert undue influence over the parameters of the model. So, if we were to delete a certain case, would we obtain different regression coefficients? This type of analysis can help to determine whether the regression model is stable across the sample, or whether it is biased by a few influential cases. Again, this process will unveil outliers.
There are several residual statistics that can be used to assess the influence of a particular case. One statistic is the adjusted predicted value for a case when that case is excluded from the analysis. In effect, the computer calculates a new model without a particular case and then uses this new model to predict the value of the outcome variable for the case that was excluded. If a case does not exert a large influence over the model then we would expect the adjusted predicted value to be very similar to the predicted value when the case is included. Put simply, if the model is stable then the predicted value of a case should be the same regardless of whether or not that case was used to calculate the model. The difference between the adjusted predicted value and the original predicted value is known as DFFit (see below). We can also look at the residual based on the adjusted predicted value: that is, the difference between the adjusted predicted value and the original observed value. This is the deleted residual. The deleted residual can be divided by the standard deviation to give a standardized value known as the Studentized deleted residual. This residual can be compared across different regression analyses because it is measured in standard units.
The deleted residuals are very useful to assess the influence of a case on the ability of the model to predict that case. However, they do not provide any information about how a case influences the model as a whole (i.e. the impact that a case has on the model’s ability to predict all cases). One statistic that does consider the effect of a single case on the model as a whole is Cook’s distance. Cook’s distance is a measure of the overall influence of a case on the model and Cook and Weisberg (1982) have suggested that values greater than 1 may be cause for concern.
A second measure of influence is leverage (sometimes called hat values), which gauges the influence of the observed value of the outcome variable over the predicted values. The average leverage value is defined as (k+1)/n in which k is the number of predictors in the model and n is the number of participants. [6] Leverage values can lie between 0 (indicating that the case has no influence whatsoever) and 1 (indicating that the case has complete influence over prediction). If no cases exert undue influence over the model then we would expect all of the leverage values to be close to the average value ((k+1)/n). Hoaglin and Welsch (1978) recommend investigating cases with values greater than twice the average (2(k+1)/n) and Stevens (2002) recommends using three times the average (3(k+1)/n) as a cut-off point for identifying cases having undue influence. We will see how to use these cut-off points later. However, cases with large leverage values will not necessarily have a large influence on the regression coefficients because they are measured on the outcome variables rather than the predictors.
Related to the leverage values are the Mahalanobis distances (Figure 7.9), which measure the distance of cases from the mean(s) of the predictor variable(s). You need to look for the cases with the highest values. It is not easy to establish a cut-off point at which to worry, although Barnett and Lewis (1978) have produced a table of critical values dependent on the number of predictors and the sample size. From their work it is clear that even with large samples (N = 500) and 5 predictors, values above 25 are cause for concern. In smaller samples (N = 100) and with fewer predictors (namely 3) values greater than 15 are problematic, and in very small samples (N = 30) with only 2 predictors values greater than 11 should be examined. However, for more specific advice, refer to Barnett and Lewis’s table.
It is possible to run the regression analysis with a case included and then rerun the analysis with that same case excluded. If we did this, undoubtedly there would be some difference between the b coefficients in the two regression equations. This difference would tell us how much influence a particular case has on the parameters of the regression model. To take a hypothetical example, imagine two variables that had a perfect negative relationship except for a single case (case 30). If a regression analysis was done on the 29 cases that were perfectly linearly related then we would get a model in which the predictor variable X perfectly predicts the outcome variable Y, and there are no errors. If we then ran the analysis but this time include the case that didn’t conform (case 30), then the resulting model has different parameters. Some data are stored in the file dfbeta.sav which illustrate such a situation. Try running a simple regression first with all the cases included and then with case 30 deleted. The results are summarized in Table 7.1, which shows: (1) the parameters for the regression model when the extreme case is included or excluded; (2) the resulting regression equations; and (3) the value of Y predicted from participant 30’s score on the X variable (which is obtained by replacing the X in the regression equation with participant 30’s score for X, which was 1).
When case 30 is excluded, these data have a perfect negative relationship; hence the coefficient for the predictor (b1) is -1 (remember that in simple regression this term is the same as Pearson’s correlation coefficient), and the coefficient for the constant (the intercept, b0) is 31. However, when case 30 is included, both parameters are reduced [7]and the difference between the parameters is also displayed. The difference between a parameter estimated using all cases and estimated when one case is excluded is known as the DFBeta in SPSS. DFBeta is calculated for every case and for each of the parameters in the model. So, in our hypothetical example, the DFBeta for the constant is -2, and the DFBeta for the predictor variable is 0.1. By looking at the values of DFBeta, it is possible to identify cases that have a large influence on the parameters of the regression model. Again, the units of measurement used will affect these values and so SPSS produces a standardized DFBeta. These standardized values are easier to use because universal cut-off points can be applied. In this case absolute values above 1 indicate cases that substantially influence the model parameters (although Stevens, 2002, suggests looking at cases with absolute values greater than 2).
A related statistic is the DFFit, which is the difference between the predicted value for a case when the model is calculated including that case and when the model is calculated excluding that case: in this example the value is -1.90 (see Table 7.1). If a case is not influential then its DFFit should be zero – hence, we expect non-influential cases to have small DFFit values. However, we have the problem that this statistic depends on the units of measurement of the outcome and so a DFFit of 0.5 will be very small if the outcome ranges from 1 to 100, but very large if the outcome varies from 0 to 1. Therefore, SPSS also produces standardized versions of the DFFit values (Standardized DFFit). A final measure is that of the covariance ratio (CVR), which is a measure of whether a case influences the variance of the regression parameters. A description of the computation of this statistic would leave most readers dazed and confused, so suffice to say that when this ratio is close to 1 the case is having very little influence on the variances of the model parameters. Belsey, Kuh and Welsch (1980) recommend the following:
• If CVRi > 1 + [3(k + 1)/n] then deleting the ith case will damage the precision of some of the model’s parameters.
• If CVRi < 1 − [3(k + 1)/n] then deleting the ith case will improve the precision of some of the model’s parameters.
In both equations, k is the number of predictors, CVRi is the covariance ratio for the ith participant, and n is the sample size.
[6] You may come across the average leverage denoted as p/n in which p is the number of parameters being estimated. In multiple regression, we estimate parameters for each predictor and also for a constant and so p is equivalent to the number of predictors plus one, (k+1).
[7] The value of b1 is reduced because the data no longer have a perfect linear relationship and so there is now variance that the model cannot explain.
7.6.1.3. A final comment on diagnostic statistics
There are a lot of diagnostic statistics that should be examined after a regression analysis, and it is difficult to summarize this wealth of material into a concise conclusion. However, one thing I would like to stress is a point made by Belsey et al. (1980) who noted the dangers inherent in these procedures. The point is that diagnostics are tools that enable you to see how good or bad your model is in terms of fitting the sampled data. They are a way of assessing your model. They are not, however, a way of justifying the removal of data points to effect some desirable change in the regression parameters (e.g. deleting a case that changes a non-significant b-value into a significant one). Stevens (2002), as ever, offers excellent advice:
If a point is a significant outlier on Y, but its Cook’s distance is < 1, there is no real need to delete that point since it does not have a large effect on the regression analysis. However, one should still be interested in studying such points further to understand why they did not fit the model. (p. 135)
7.6.2.1. Checking assumptions
To draw conclusions about a population based on a regression analysis done on a sample, several assumptions must be true (see Berry, 1993):
Variable types: All predictor variables must be quantitative or categorical (with two categories), and the outcome variable must be quantitative, continuous and unbounded. By quantitative I mean that they should be measured at the interval level and by unbounded I mean that there should be no constraints on the variability of the outcome. If the outcome is a measure ranging from 1 to 10 yet the data collected vary between 3 and 7, then these data are constrained.
Non-zero variance: The predictors should have some variation in value (i.e. they do not have variances of 0).
No perfect multicollinearity: There should be no perfect linear relationship between two or more of the predictors. So, the predictor variables should not correlate too highly (see section 7.6.2.4).
Predictors are uncorrelated with ‘external variables’: External variables are variables that haven’t been included in the regression model which influence the outcome variable. [8] These variables can be thought of as similar to the ‘third variable’ that was discussed with reference to correlation. This assumption means that there should be no external variables that correlate with any of the variables included in the regression model. Obviously, if external variables do correlate with the predictors, then the conclusions we draw from the model become unreliable (because other variables exist that can predict the outcome just as well).
Homoscedasticity: At each level of the predictor variable(s), the variance of the residual terms should be constant. This just means that the residuals at each level of the predictor(s) should have the same variance (homoscedasticity); when the variances are very unequal there is said to be heteroscedasticity (see section 5.6 as well).
Independent errors: For any two observations the residual terms should be uncorrelated (or independent). This eventuality is sometimes described as a lack of autocorrelation. This assumption can be tested with the Durbin–Watson test, which tests for serial correlations between errors. Specifically, it tests whether adjacent residuals are correlated. The test statistic can vary between 0 and 4 with a value of 2 meaning that the residuals are uncorrelated. A value greater than 2 indicates a negative correlation between adjacent residuals, whereas a value below 2 indicates a positive correlation. The size of the Durbin–Watson statistic depends upon the number of predictors in the model and the number of observations. For accuracy, you should look up the exact acceptable values in Durbin and Watson’s (1951) original paper. As a very conservative rule of thumb, values less than 1 or greater than 3 are definitely cause for concern; however, values closer to 2 may still be problematic depending on your sample and model.
Normally distributed errors: It is assumed that the residuals in the model are random, normally distributed variables with a mean of 0. This assumption simply means that the differences between the model and the observed data are most frequently zero or very close to zero, and that differences much greater than zero happen only occasionally. Some people confuse this assumption with the idea that predictors have to be normally distributed. In fact, predictors do not need to be normally distributed (see section 7.11).
Independence: It is assumed that all of the values of the outcome variable are independent (in other words, each value of the outcome variable comes from a separate entity).
Linearity: The mean values of the outcome variable for each increment of the predictor(s) lie along a straight line. In plain English this means that it is assumed that the relationship we are modelling is a linear one. If we model a non-linear relationship using a linear model then this obviously limits the generalizability of the findings.
This list of assumptions probably seems pretty daunting but as we saw in Chapter 5, assumptions are important. When the assumptions of regression are met, the model that we get for a sample can be accurately applied to the population of interest (the coefficients and parameters of the regression equation are said to be unbiased). Some people assume that this means that when the assumptions are met the regression model from a sample is always identical to the model that would have been obtained had we been able to test the entire population. Unfortunately, this belief isn’t true. What an unbiased model does tell us is that on average the regression model from the sample is the same as the population model. However, you should be clear that even when the assumptions are met, it is possible that a model obtained from a sample may not be the same as the population model – but the likelihood of them being the same is increased.
[8] Some authors choose to refer to these external variables as part of an error term that includes any random factor in the way in which the outcome varies. However, to avoid confusion with the residual terms in the regression equations I have chosen the label ‘external variables’. Although this term implicitly washes over any random factors, I acknowledge their presence here!
7.6.2.4. Multicollinearity
Multicollinearity exists when there is a strong correlation between two or more predictors in a regression model. Multicollinearity poses a problem only for multiple regression because (without wishing to state the obvious) simple regression requires only one predictor. Perfect collinearity exists when at least one predictor is a perfect linear combination of the others (the simplest example being two predictors that are perfectly correlated – they have a correlation coefficient of 1). If there is perfect collinearity between predictors it becomes impossible to obtain unique estimates of the regression coefficients because there are an infinite number of combinations of coefficients that would work equally well. Put simply, if we have two predictors that are perfectly correlated, then the values of b for each variable are interchangeable. The good news is that perfect collinearity is rare in real-life data. The bad news is that less than perfect collinearity is virtually unavoidable. Low levels of collinearity pose little threat to the models generated by SPSS, but as collinearity increases there are three problems that arise:
Untrustworthy bs: As collinearity increases so do the standard errors of the b coefficients. If you think back to what the standard error represents, then big standard errors for b coefficients means that these bs are more variable across samples. Therefore, it means that the b coefficient in our sample is less likely to represent the population. Crudely put, multicollinearity means that the b-values are less trustworthy. Don’t lend them money and don’t let them go for dinner with your boy- or girlfriend. Of course if the bs are variable from sample to sample then the resulting predictor equations will be unstable across samples too.
It limits the size of R: Remember that R is a measure of the multiple correlation between the predictors and the outcome and that R² indicates the variance in the outcome for which the predictors account. Imagine a situation in which a single variable predicts the outcome variable fairly successfully (e.g. R = .80) and a second predictor variable is then added to the model. This second variable might account for a lot of the variance in the outcome (which is why it is included in the model), but the variance it accounts for is the same variance accounted for by the first variable. In other words, once the variance accounted for by the first predictor has been removed, the second predictor accounts for very little of the remaining variance (the second variable accounts for very little unique variance). Hence, the overall variance in the outcome accounted for by the two predictors is little more than when only one predictor is used (so R might increase from .80 to .82). This idea is connected to the notion of partial correlation that was explained in Chapter 6. If, however, the two predictors are completely uncorrelated, then the second predictor is likely to account for different variance in the outcome to that accounted for by the first predictor. So, although in itself the second predictor might account for only a little of the variance in the outcome, the variance it does account for is different to that of the other predictor (and so when both predictors are included, R is substantially larger, say .95). Therefore, having uncorrelated predictors is beneficial.
Importance of predictors: Multicollinearity between predictors makes it difficult to assess the individual importance of a predictor. If the predictors are highly correlated, and each accounts for similar variance in the outcome, then how can we know which of the two variables is important? Quite simply we can’t tell which variable is important – the model could include either one, interchangeably.
One way of identifying multicollinearity is to scan a correlation matrix of all of the predictor variables and see if any correlate very highly (by very highly I mean correlations of above .80 or .90). This is a good ‘ball park’ method but misses more subtle forms of multicollinearity. Luckily, SPSS produces various collinearity diagnostics, one of which is the variance inflation factor (VIF). The VIF indicates whether a predictor has a strong linear relationship with the other predictor(s). Although there are no hard and fast rules about what value of the VIF should cause concern, Myers (1990) suggests that a value of 10 is a good value at which to worry. What’s more, if the average VIF is greater than 1, then multicollinearity may be biasing the regression model (Bowerman & O’Connell, 1990). Related to the VIF is the tolerance statistic, which is its reciprocal (1/VIF). As such, values below 0.1 indicate serious problems although Menard (1995) suggests that values below 0.2 are worthy of concern.
Other measures that are useful in discovering whether predictors are dependent are the eigenvalues of the scaled, uncentred cross-products matrix, the condition indexes and the variance proportions. These statistics are extremely complex and will be covered as part of the interpretation of SPSS output (see section 7.8.5). If none of this has made any sense then have a look at Hutcheson and Sofroniou (1999: 78–85) who give a really clear explanation of multicollinearity.
7.8. Interpreting multiple regression
7.8.2. Summary of model
The next section of output describes the overall model (so it tells us whether the model is successful in predicting record sales). Remember that we chose a hierarchical method and so each set of summary statistics is repeated for each stage in the hierarchy. In SPSS Output 7.5 you should note that there are two models. Model 1 refers to the first stage in the hierarchy when only advertising budget is used as a predictor. Model 2 refers to when all three predictors are used. SPSS Output 7.5 is the model summary and this table was produced using the Model fit option. This option is selected by default in SPSS because it provides us with some very important information about the model: the values of R, R² and the adjusted R². If the R squared change and Durbin-Watson options were selected, then these values are included also (if they weren’t selected you’ll find that you have a smaller table).
The model summary table is shown in SPSS Output 7.5 and you should notice that under this table SPSS tells us what the dependent variable (outcome) was and what the predictors were in each of the two models. In the column labelled R are the values of the multiple correlation coefficient between the predictors and the outcome. When only advertising budget is used as a predictor, this is the simple correlation between advertising and record sales (0.578). In fact all of the statistics for model 1 are the same as the simple regression model earlier (see section 7.4). The next column gives us a value of R², which we already know is a measure of how much of the variability in the outcome is accounted for by the predictors. For the first model its value is .335, which means that advertising budget accounts for 33.5% of the variation in record sales. However, when the other two predictors are included as well (model 2), this value increases to .665 or 66.5% of the variance in record sales. Therefore, if advertising accounts for 33.5%, we can tell that attractiveness and radio play account for an additional 33%. [9] So, the inclusion of the two new predictors has explained quite a large amount of the variation in record sales.
The adjusted R² gives us some idea of how well our model generalizes and ideally we would like its value to be the same, or very close to, the value of R². In this example the difference for the final model is small (in fact the difference between the values is .665 – .660 = .005 (about 0.5%). This shrinkage means that if the model were derived from the population rather than a sample it would account for approximately 0.5% less variance in the outcome. Advanced students might like to apply Stein’s formula to the R² to get some idea of its likely value in different samples. Stein’s formula was given in equation (7.11) and can be applied by replacing n with the sample size (200) and k with the number of predictors (3):
adjusted R² = 1 – [(200-1/200-3-1)(200-2/200-3-2)(200+1/200)] (1-0.665)
=1 – [(1.015)(1.015)(1.005)] (0.335)
= 1 – 0.347
= 0.653
This value is very similar to the observed value of R² (.665) indicating that the cross-validity of this model is very good.
The change statistics are provided only if requested and these tell us whether the change in R² is significant. The significance of R² can actually be tested using an F-ratio, and this F is calculated from the following equation (in which N is the number of cases or participants, and k is the number of predictors in the model):
F = (N−k−1)R² / k(1−R²)
In SPSS Output 7.5, the change in this F is reported for each block of the hierarchy. So, model 1 causes R² to change from 0 to .335, and this change in the amount of variance explained gives rise to an F-ratio of 99.587, which is significant with a probability less than .001. Bearing in mind for this first model that we have only one predictor (so k = 1) and 200 cases (N = 200), this F comes from the equation above: [10]
FModel1 = ((200-1-1)*0.334648) / (1*(1-0.334648)) = 99.587
The addition of the new predictors (model 2) causes R² to increase by .330 (see above). We can calculate the F-ratio for this change using the same equation, but because we’re looking at the change in models we use the change in R², R²Change, and the R² in the new model (model 2 in this case so I’ve called it R22) and we also use the change in the number of predictors, kChange (model 1 had one predictor and model 2 had three predictors, so the change in the number of predictors is 3-1 = 2), and the number of predictors in the new model, k2 (in this case because we’re looking at model 2). Again, if we use a few more decimal places than in the SPSS table, we get approximately the same answer as SPSS:
FChange = (N – k2 – 1) R²Change / kChange (1 – R22)
= ((200 – 3 – 1) × 0.330) / (2*(1 – 0.664668))
= 96.44
As such, the change in the amount of variance that can be explained gives rise to an F-ratio of 96.44, which is again significant (p < .001). The change statistics therefore tell us about the difference made by adding new predictors to the model.
Finally, if you requested the Durbin–Watson statistic it will be found in the last column of the table in SPSS Output 7.5. This statistic informs us about whether the assumption of independent errors is tenable (see section 7.6.2.1). As a conservative rule I suggested that values less than 1 or greater than 3 should definitely raise alarm bells (although I urge you to look up precise values for the situation of interest). The closer to 2 that the value is, the better, and for these data the value is 1.950, which is so close to 2 that the assumption has almost certainly been met.
SPSS Output 7.6 shows the next part of the output, which contains an ANOVA that tests whether the model is significantly better at predicting the outcome than using the mean as a ‘best guess’. Specifically, the F-ratio represents the ratio of the improvement in prediction that results from fitting the model, relative to the inaccuracy that still exists in the model (see section 7.2.3). This table is again split into two sections: one for each model. We are told the value of the sum of squares for the model (this value is SSM in section 7.2.3 and represents the improvement in prediction resulting from fitting a regression line to the data rather than using the mean as an estimate of the outcome). We are also told the residual sum of squares (this value is SSR in section 7.2.3 and represents the total difference between the model and the observed data). We are also told the degrees of freedom (df) for each term. In the case of the improvement due to the model, this value is equal to the number of predictors (1 for the first model and 3 for the second), and for SSR it is the number of observations (200) minus the number of coefficients in the regression model. The first model has two coefficients (one for the predictor and one for the constant) whereas the second has four (one for each of the three predictors and one for the constant). Therefore, model 1 has 198 degrees of freedom whereas model 2 has 196. The average sum of squares (MS) is then calculated for each term by dividing the SS by the df. The F-ratio is calculated by dividing the average improvement in prediction by the model (MSM) by the average difference between the model and the observed data (MSR). If the improvement due to fitting the regression model is much greater than the inaccuracy within the model then the value of F will be greater than 1 and SPSS calculates the exact probability of obtaining the value of F by chance. For the initial model the F-ratio is 99.587, which is very unlikely to have happened by chance (p < .001). For the second model the value of F is even higher (129.498), which is also highly significant (p < .001). We can interpret these results as meaning that the initial model significantly improved our ability to predict the outcome variable, but that the new model (with the extra predictors) was even better (because the F-ratio is more significant).
[9] That is, 33% = 66.5% − 33.5% (this value is the R Square Change in the table).
[10] To get the same values as SPSS we have to use the exact value of R², which is 0.3346480676231 (if you don’t believe me double-click on the table in the SPSS output that reports this value, then double-click on the cell of the table containing the value of R² and you’ll see that .335 becomes the value that I’ve just typed!).
7.8.3. Model parameters
Remember that in multiple regression the model takes the form of equation (7.9) and in that equation there are several unknown quantities (the b-values). The first part of the table gives us estimates for these b-values and these values indicate the individual contribution of each predictor to the model. If we replace the b-values in equation (7.9) we find that we can define the model as follows:
salesi = b0 + b1advertisingi + b2airplayi + b3attractivenessi = − 26:61 + (0.08advertisingi) + (3.37airplayi) + (11.09attractivenessi)
Advertising budget (b = 0.085): This value indicates that as advertising budget increases by one unit, record sales increase by 0.085 units. Both variables were measured in thousands; therefore, for every £1000 more spent on advertising, an extra 0.085 thousand records (85 records) are sold. This interpretation is true only if the effects of attractiveness of the band and airplay are held constant.
Advertising budget (standardized β = .511): This value indicates that as advertising budget increases by one standard deviation (£485,655), record sales increase by 0.511 standard deviations. The standard deviation for record sales is 80,699 and so this constitutes a change of 41,240 sales (0.511 × 80,699). Therefore, for every £485,655 more spent on advertising, an extra 41,240 records are sold. This interpretation is true only if the effects of attractiveness of the band and airplay are held constant.
7.8.5. Assessing the assumption of no multicollinearity
… SPSS also produces a table of eigenvalues of the scaled, uncentred cross-products matrix, condition indexes and variance proportions (see Jane Superbrain Box 7.2). There is a lengthy discussion, and example, of collinearity in section 8.8.1 and how to detect it using variance proportions, so I will limit myself now to saying that we are looking for large variance proportions on the same small eigenvalues. Therefore, in SPSS Output 7.9 we look at the bottom few rows of the table (these are the small eigenvalues) and look for any variables that both have high variance proportions for that eigenvalue. The variance proportions vary between 0 and 1, and for each predictor should be distributed across different dimensions (or eigenvalues). For this model, you can see that each predictor has most of its variance loading onto a different dimension (advertising has 96% of variance on dimension 2, airplay has 93% of variance on dimension 3 and attractiveness has 92% of variance on dimension 4). These data represent a classic example of no multicollinearity. For an example of when collinearity exists in the data and some suggestions about what can be done, see Chapters 8 (section 8.8.1) and 17 (section 17.3.3.3).
JANE SUPERBRAIN 7.2 What are eigenvectors and eigenvalues?
The definitions and mathematics of eigenvalues and eigenvectors are very complicated and most of us need not worry about them (although they do crop up again in Chapters 16 and 17). However, although the mathematics of them is hard, they are quite easy to visualize! Imagine we have two variables: the salary a supermodel earns in a year, and how attractive she is. Also imagine these two variables are normally distributed and so can be considered together as a bivariate normal distribution. If these variables are correlated, then their scatterplot forms an ellipse. This is shown in the scatterplots below: if we draw a dashed line around the outer values of the scatterplot we get something oval shaped. Now, we can draw two lines to measure the length and height of this ellipse. These lines are the eigenvectors of the original correlation matrix for these two variables (a vector is just a set of numbers that tells us the location of a line in geometric space). Note that the two lines we’ve drawn (one for height and one for width of the oval) are perpendicular; that is, they are at 90 degrees, which means that they are independent of one another). So, with two variables, eigenvectors are just lines measuring the length and height of the ellipse that surrounds the scatterplot of data for those variables. If we add a third variable (e.g. experience of the supermodel) then all that happens is our scatterplot gets a third dimension, the ellipse turns into something shaped like a rugby ball (or American football), and because we now have a third dimension (height, width and depth) we get an extra eigenvector to measure this extra dimension. If we add a fourth variable, a similar logic applies (although it’s harder to visualize): we get an extra dimension, and an eigenvector to measure that dimension. Now, each eigenvector has an eigenvalue that tells us its length (i.e. the distance from one end of the eigenvector to the other). So, by looking at all of the eigenvalues for a data set, we know the dimensions of the ellipse or rugby ball: put more generally, we know the dimensions of the data. Therefore, the eigenvalues show how evenly (or otherwise) the variances of the matrix are distributed.
In the case of two variables, the condition of the data is related to the ratio of the larger eigenvalue to the smaller. Let’s look at the two extremes: when there is no relationship at all between variables, and when there is a perfect relationship. When there is no relationship, the scatterplot will, more or less, be contained within a circle (or a sphere if we had three variables). If we again draw lines that measure the height and width of this circle we’ll find that these lines are the same length. The eigenvalues measure the length, therefore the eigenvalues will also be the same. So, when we divide the largest eigenvalue by the smallest we’ll get a value of 1 (because the eigenvalues are the same). When the variables are perfectly correlated (i.e. there is perfect collinearity) then the scatterplot forms a straight line and the ellipse surrounding it will also collapse to a straight line. Therefore, the height of the ellipse will be very small indeed (it will approach zero). Therefore, when we divide the largest eigenvalue by the smallest we’ll get a value that tends to infinity (because the smallest eigenvalue is close to zero). Therefore, an infinite condition index is a sign of deep trouble.
7.8.6. Casewise diagnostics
SPSS produces a summary table of the residual statistics and these should be examined for extreme cases. SPSS Output 7.10 shows any cases that have a standardized residual less than -2 or greater than 2 (remember that we changed the default criterion from 3 to 2 in Figure 7.14). I mentioned in section 7.6.1.1 that in an ordinary sample we would expect 95% of cases to have standardized residuals within about ±2. We have a sample of 200, therefore it is reasonable to expect about 10 cases (5%) to have standardized residuals outside of these limits. From SPSS Output 7.10 we can see that we have 12 cases (6%) that are outside of the limits: therefore, our sample is within 1% of what we would expect. In addition, 99% of cases should lie within ±2.5 and so we would expect only 1% of cases to lie outside of these limits. From the cases listed here, it is clear that two cases (1%) lie outside of the limits (cases 164 and 169). Therefore, our sample appears to conform to what we would expect for a fairly accurate model. These diagnostics give us no real cause for concern except that case 169 has a standardized residual greater than 3, which is probably large enough for us to investigate this case further. [...]
SPSS Output 7.11 shows the influence statistics for the 12 cases that I selected. None of them have a Cook’s distance greater than 1 (even case 169 is well below this criterion) and so none of the cases is having an undue influence on the model. The average leverage can be calculated as 0.02 (k+1/n = 4/200) and so we are looking for values either twice as large as this (0.04) or three times as large (0.06) depending on which statistician you trust most (see section 7.6.1.2)! All cases are within the boundary of three times the average and only case 1 is close to two times the average. Finally, from our guidelines for the Mahalanobis distance we saw that with a sample of 100 and three predictors, values greater than 15 were problematic. We have three predictors and a larger sample size, so this value will be a conservative cut-off, yet none of our cases comes close to exceeding this criterion. The evidence suggests that there are no influential cases within our data (although all cases would need to be examined to confirm this fact).
We can look also at the DFBeta statistics to see whether any case would have a large influence on the regression parameters. An absolute value greater than 1 is a problem and in all cases the values lie within ±1, which shows that these cases have no undue influence over the regression parameters. There is also a column for the covariance ratio. We saw in section 7.6.1.2 that we need to use the following criteria:
• CVRi > 1 + [3(k + 1)/n] = 1 + [3(3 + 1)/200] = 1.06.
• CVRi < 1 – [3(k + 1)/n] = 1 – [3(3 + 1)/200] = 0.94.
Therefore, we are looking for any cases that deviate substantially from these boundaries. Most of our 12 potential outliers have CVR values within or just outside these boundaries. The only case that causes concern is case 169 (again!) whose CVR is some way below the bottom limit. However, given the Cook’s distance for this case, there is probably little cause for alarm.
CRAMMING SAM’s Tips
You need to look for cases that might be influencing the regression model:
• Look at standardized residuals and check that no more than 5% of cases have absolute values above 2, and that no more than about 1% have absolute values above 2.5. Any case with the value above about 3, could be an outlier.
• Look in the data editor for the values of Cook’s distance: any value above 1 indicates a case that might be influencing the model.
• Calculate the average leverage (the number of predictors plus 1, divided by the sample size) and then look for values greater than twice or three times this average value.
• For Mahalanobis distance, a crude check is to look for values above 25 in large samples (500) and values above 15 in smaller samples (100). However, Barnett and Lewis (1978) should be consulted for more detailed analysis.
• Look for absolute values of DFBeta greater than 1.
• Calculate the upper and lower limit of acceptable values for the covariance ratio, CVR. The upper limit is 1 plus three times the average leverage, whereas the lower limit is 1 minus three times the average leverage. Cases that have a CVR that fall outside of these limits may be problematic.
7.8.7. Checking assumptions
As a final stage in the analysis, you should check the assumptions of the model. We have already looked for collinearity within the data and used Durbin–Watson to check whether the residuals in the model are independent. In section 7.7.4 we asked for a plot of *ZRESID against *ZPRED and for a histogram and normal probability plot of the residuals. The graph of *ZRESID and *ZPRED should look like a random array of dots evenly dispersed around zero. If this graph funnels out, then the chances are that there is heteroscedasticity in the data. If there is any sort of curve in this graph then the chances are that the data have broken the assumption of linearity. Figure 7.19 shows several examples of the plot of standardized residuals against standardized predicted values. Panel (a) shows the graph for the data in our record sales example. Note how the points are randomly and evenly dispersed throughout the plot. This pattern is indicative of a situation in which the assumptions of linearity and homoscedasticity have been met. Panel (b) shows a similar plot for a data set that violates the assumption of homoscedasticity. Note that the points form the shape of a funnel so they become more spread out across the graph. This funnel shape is typical of heteroscedasticity and indicates increasing variance across the residuals. Panel (c) shows a plot of some data in which there is a non-linear relationship between the outcome and the predictor. This pattern is shown up by the residuals. A line illustrating the curvilinear relationship has been drawn over the top of the graph to illustrate the trend in the data. Finally, panel (d) represents a situation in which the data not only represent a non-linear relationship, but also show heteroscedasticity. Note first the curved trend in the data, and then also note that at one end of the plot the points are very close together whereas at the other end they are widely dispersed. When these assumptions have been violated you will not see these exact patterns, but hopefully these plots will help you to understand the types of anomalies you should look out for.
To test the normality of residuals, we must look at the histogram and normal probability plot selected in Figure 7.15. Figure 7.20 shows the histogram and normal probability plot of the data for the current example (left-hand side). The histogram should look like a normal distribution (a bell-shaped curve). SPSS draws a curve on the histogram to show the shape of the distribution. For the record company data, the distribution is roughly normal (although there is a slight deficiency of residuals exactly on zero). Compare this histogram to the extremely non-normal histogram next to it and it should be clear that the non-normal distribution is extremely skewed (unsymmetrical). So, you should look for a curve that has the same shape as the one for the record sales data: any deviation from this curve is a sign of non-normality and the greater the deviation, the more non-normally distributed the residuals. The normal probability plot also shows up deviations from normality (see Chapter 5). The straight line in this plot represents a normal distribution, and the points represent the observed residuals. Therefore, in a perfectly normally distributed data set, all points will lie on the line. This is pretty much what we see for the record sales data. However, next to the normal probability plot of the record sales data is an example of an extreme deviation from normality. In this plot, the dots are very distant from the line, which indicates a large deviation from normality. For both plots, the non-normal data are extreme cases and you should be aware that the deviations from normality are likely to be subtler. Of course, you can use what you learnt in Chapter 5 to do a K–S test on the standardized residuals to see whether they deviate significantly from normality.
A final set of plots specified in Figure 7.15 was the partial plots. These plots are scatterplots of the residuals of the outcome variable and each of the predictors when both variables are regressed separately on the remaining predictors. I mentioned earlier that obvious outliers on a partial plot represent cases that might have undue influence on a predictor’s regression coefficient and that non-linear relationships and heteroscedasticity can be detected using these plots as well:
CRAMMING SAM’s Tips
You need to check some of the assumptions of regression to make sure your model generalizes beyond your sample:
• Look at the graph of *ZRESID plotted against *ZPRED. If it looks like a random array of dots then this is good. If the dots seem to get more or less spread out over the graph (look like a funnel) then this is probably a violation of the assumption of homogeneity of variance. If the dots have a pattern to them (i.e. a curved shape) then this is probably a violation of the assumption of linearity. If the dots seem to have a pattern and are more spread out at some points on the plot than others then this probably reflects violations of both homogeneity of variance and linearity. Any of these scenarios puts the validity of your model into question. Repeat the above for all partial plots too.
• Look at histograms and P–P plots. If the histograms look like normal distributions (and the P–P plot looks like a diagonal line), then all is well. If the histogram looks non-normal and the P–P plot looks like a wiggly snake curving around a diagonal line then things are less good! Be warned, though: distributions can look very non-normal in small samples even when they are!
7.9. What if I violate an assumption?
It’s worth remembering that you can have a perfectly good model for your data (no outliers, influential cases, etc.) and you can use that model to draw conclusions about your sample, even if your assumptions are violated. However, it’s much more interesting to generalize your regression model and this is where assumptions become important. If they have been violated then you cannot generalize your findings beyond your sample. The options for correcting for violated assumptions are a bit limited. If residuals show problems with heteroscedasticity or non-normality you could try transforming the raw data – but this won’t necessarily affect the residuals! If you have a violation of the linearity assumption then you could see whether you can do logistic regression instead (described in the next chapter). Finally, there are a series of robust regression techniques (see section 5.7.4), which are described extremely well by Rand Wilcox in Chapter 10 of his book. SPSS can’t do these methods directly (well, technically it can do robust parameter estimates but it’s not easy), but you can attempt a robust regression using some of Wilcox’s files for the software R (Wilcox, 2005) using SPSS’s R plugin.
7.11. Categorical predictors and multiple regression
Often in regression analysis you’ll collect data about groups of people (e.g. ethnic group, gender, socio-economic status, diagnostic category). You might want to include these groups as predictors in the regression model; however, we saw from our assumptions that variables need to be continuous or categorical with only two categories. We saw in section 6.5.5 that a point–biserial correlation is Pearson’s r between two variables when one is continuous and the other has two categories coded as 0 and 1. We’ve also learnt that simple regression is based on Pearson’s r, so it shouldn’t take a great deal of imagination to see that, like the point–biserial correlation, we could construct a regression model with a predictor that has two categories (e.g. gender). Likewise, it shouldn’t be too inconceivable that we could then extend this model to incorporate several predictors that had two categories. All that is important is that we code the two categories with the values of 0 and 1. Why is it important that there are only two categories and that they’re coded 0 and 1? Actually, I don’t want to get into this here because this chapter is already too long, the publishers are going to break my legs if it gets any longer, and I explain it anyway later in the book (sections 9.7 and 10.2.3) so, for the time being, just trust me!
Chapter 8: Logistic regression
8.3.1. Assessing the model: the log-likelihood statistic
We’ve seen that the logistic regression model predicts the probability of an event occurring for a given person (we would denote this as P(Yi) the probability that Y occurs for the ith person), based on observations of whether or not the event did occur for that person (we could denote this as Yi, the actual outcome for the ith person). So, for a given person, Y will be either 0 (the outcome didn’t occur) or 1 (the outcome did occur), and the predicted value, P(Y), will be a value between 0 (there is no chance that the outcome will occur) and 1 (the outcome will certainly occur). We saw in multiple regression that if we want to assess whether a model fits the data we can compare the observed and predicted values of the outcome (if you remember, we use R², which is the Pearson correlation between observed values of the outcome and the values predicted by the regression model). Likewise, in logistic regression, we can use the observed and predicted values to assess the fit of the model. The measure we use is the log-likelihood:
log-likelihood = ΣN i = 1 [Yi ln(P(Yi)) + (1-Yi) ln(1-P(Yi))] (8.5)
The log-likelihood is based on summing the probabilities associated with the predicted and actual outcomes (Tabachnick & Fidell, 2007). The log-likelihood statistic is analogous to the residual sum of squares in multiple regression in the sense that it is an indicator of how much unexplained information there is after the model has been fitted. It, therefore, follows that large values of the log-likelihood statistic indicate poorly fitting statistical models, because the larger the value of the log-likelihood, the more unexplained observations there are.
Now, it’s possible to calculate a log-likelihood for different models and to compare these models by looking at the difference between their log-likelihoods. One use of this is to compare the state of a logistic regression model against some kind of baseline state. The baseline state that’s usually used is the model when only the constant is included. In multiple regression, the baseline model we use is the mean of all scores (this is our best guess of the outcome when we have no other information). In logistic regression, if we want to predict the outcome, what would our best guess be? Well, we can’t use the mean score because our outcome is made of zeros and ones and so the mean is meaningless! However, if we know the frequency of zeros and ones, then the best guess will be the category with the largest number of cases. So, if the outcome occurs 107 times, and doesn’t occur only 72 times, then our best guess of the outcome will be that it occurs (because it occurs 107 times compared to only 72 times when it doesn’t occur). As such, like multiple regression, our baseline model is the model that gives us the best prediction when we know nothing other than the values of the outcome: in logistic regression this will be to predict the outcome that occurs most often. This is, the logistic regression model when only the constant is included. If we then add one or more predictors to the model, we can compute the improvement of the model as follows:
χ² = 2 [LL(new)−LL(baseline)]
(df = knew − kbaseline) (8.6)
So, we merely take the new model and subtract from it the baseline model (the model when only the constant is included). You’ll notice that we multiply this value by 2; this is because it gives the result a chi-square distribution (see Chapter 18 and the Appendix) and so makes it easy to calculate the significance of the value. The chi-square distribution we use has degrees of freedom equal to the number of parameters, k in the new model minus the number of parameters in the baseline model. The number of parameters in the baseline model will always be 1 (the constant is the only parameter to be estimated); any subsequent model will have degrees of freedom equal to the number of predictors plus 1 (i.e. the number of predictors plus one parameter representing the constant).
8.3.3. Assessing the contribution of predictors: the Wald statistic
As in linear regression, we want to know not only how well the model overall fits the data, but also the individual contribution of predictors. In linear regression, we used the estimated regression coefficients (b) and their standard errors to compute a t-statistic. In logistic regression there is an analogous statistic known as the Wald statistic, which has a special distribution known as the chi-square distribution. Like the t-test in linear regression, the Wald statistic tells us whether the b coefficient for that predictor is significantly different from zero. If the coefficient is significantly different from zero then we can assume that the predictor is making a significant contribution to the prediction of the outcome (Y):
Wald = b/SEb (8.11)
Equation (8.11) shows how the Wald statistic is calculated and you can see it’s basically identical to the t-statistic in linear regression (see equation (7.6)): it is the value of the regression coefficient divided by its associated standard error. The Wald statistic (Figure 8.2) is usually used to ascertain whether a variable is a significant predictor of the outcome; however, it is probably more accurate to examine the likelihood ratio statistics. The reason why the Wald statistic should be used cautiously is because, when the regression coefficient (b) is large, the standard error tends to become inflated, resulting in the Wald statistic being underestimated (see Menard, 1995). The inflation of the standard error increases the probability of rejecting a predictor as being significant when in reality it is making a significant contribution to the model (i.e. you are more likely to make a Type II error).
8.3.4. The odds ratio: Exp(B)
More crucial to the interpretation of logistic regression is the value of the odds ratio (Exp(B) in the SPSS output), which is an indicator of the change in odds resulting from a unit change in the predictor. As such, it is similar to the b coefficient in logistic regression but easier to understand (because it doesn’t require a logarithmic transformation). When the predictor variable is categorical the odds ratio is easier to explain, so imagine we had a simple example in which we were trying to predict whether or not someone got pregnant from whether or not they used a condom last time they made love. The odds of an event occurring are defined as the probability of an event occurring divided by the probability of that event not occurring (see equation (8.12)) and should not be confused with the more colloquial usage of the word to refer to probability. So, for example, the odds of becoming pregnant are the probability of becoming pregnant divided by the probability of not becoming pregnant:
odds = P(event)/P(no event) (8.12)
P(event Y) = 1/(1 + e – (b0 + b1X1))
P(no event Y) = 1 – P(event Y)
To calculate the change in odds that results from a unit change in the predictor, we must first calculate the odds of becoming pregnant given that a condom wasn’t used. We then calculate the odds of becoming pregnant given that a condom was used. Finally, we calculate the proportionate change in these two odds.
To calculate the first set of odds, we need to use equation (8.3) to calculate the probability of becoming pregnant given that a condom wasn’t used. If we had more than one predictor we would use equation (8.4). There are three unknown quantities in this equation: the coefficient of the constant (b0), the coefficient for the predictor (b1) and the value of the predictor itself (X). We’ll know the value of X from how we coded the condom use variable (chances are we would’ve used 0 = condom wasn’t used and 1 = condom was used). The values of b1 and b0 will be estimated for us. We can calculate the odds as in equation (8.12).
Next, we calculate the same thing after the predictor variable has changed by one unit. In this case, because the predictor variable is dichotomous, we need to calculate the odds of getting pregnant, given that a condom was used. So, the value of X is now 1 (rather than 0).
We now know the odds before and after a unit change in the predictor variable. It is a simple matter to calculate the proportionate change in odds by dividing the odds after a unit change in the predictor by the odds before that change:
∆odds = odds after a unit change in the predictor/original odds (8.13)
This proportionate change in odds is the odds ratio, and we can interpret it in terms of the change in odds: if the value is greater than 1 then it indicates that as the predictor increases, the odds of the outcome occurring increase. Conversely, a value less than 1 indicates that as the predictor increases, the odds of the outcome occurring decrease. We’ll see how this works with a real example shortly.
SPSS TIP 8.1 Error messages about ‘failure to coverage’
Many statistical procedures use an iterative process, which means that SPSS attempts to estimate the parameters of the model by finding successive approximations of those parameters. Essentially, it starts by estimating the parameters with a ‘best guess’. It then attempts to approximate them more accurately (known as an iteration). It then tries again, and then again, and so on through many iterations. It stops either when the approximations of parameters converge (i.e. at each new attempt the ‘approximations’ of parameters are the same or very similar to the previous attempt), or when it reaches the maximum number of attempts (iterations).
Sometimes you will get an error message in the output that says something like ‘Maximum number of iterations were exceeded, and the log-likelihood value and/or the parameter estimates cannot converge’. What this means is that SPSS has attempted to estimate the parameters the maximum number of times (as specified in the options) but they are not converging (i.e. at each iteration SPSS is getting quite different estimates). This certainly means that you should ignore any output that SPSS has produced, and it might mean that your data are beyond help. You can try increasing the number of iterations that SPSS attempts, or make the criteria that SPSS uses to assess ‘convergence’ less strict.
8.4.4. Overdispersion
I’m not a statistician, and most of what I’ve read on overdispersion doesn’t make an awful lot of sense to me. From what I can gather, it is when the observed variance is bigger than expected from the logistic regression model. This can happen for two reasons. The first is correlated observations (i.e. when the assumption of independence is broken) and the second is due to variability in success probabilities. For example, imagine our outcome was whether a puppy in a litter survived or died. Genetic factors mean that within a given litter the chances of success (living) depend on the litter from which the puppy came. As such success probabilities vary across litters (Halekoh & Højsgaard, 2007), this example of dead puppies is particularly good – not because I’m a cat lover, but because it shows how variability in success probabilities can create correlation between observations (the survival rates of puppies from the same litter are not independent).
Overdispersion creates a problem because it tends to limit standard errors and result in narrower confidence intervals for test statistics of predictors in the logistic regression model. Given that the test statistics are computed by dividing by the standard error, if the standard error is too small then the test statistic will be bigger than it should be, and more likely to be deemed significant. Similarly, narrow confidence intervals will give us overconfidence in the effect of our predictors on the outcome. In short, there is more chance of Type I errors. The parameters themselves (i.e. the b-values) are unaffected.
SPSS produces a chi-square goodness-of-fit statistic, and overdispersion is present if the ratio of this statistic to its degrees of freedom is greater than 1 (this ratio is called the dispersion parameter, φ). Overdispersion is likely to be problematic if the dispersion parameter approaches or is greater than 2. (Incidentally, underdispersion is shown by values less than 1, but this problem is much less common in practice.) There is also the deviance goodness-of-fit statistic, and the dispersion parameter can be based on this statistic instead (again by dividing by the degrees of freedom). When the chi-square and deviance statistics are very discrepant, then overdispersion is likely.
The effects of overdispersion can be reduced by using the dispersion parameter to rescale the standard errors and confidence intervals. For example, the standard errors are multiplied by √φ to make them bigger (as a function of how big the overdispersion is). You can base these corrections on the deviance statistic too, and whether you rescale using this statistic or the Pearson chi-square statistic depends on which one is bigger. The bigger statistic will have the bigger dispersion parameter (because their degrees of freedom are the same), and will make the bigger correction; therefore, correct by the bigger of the two.
8.5.4. Obtaining residuals
As with linear regression, it is possible to save a set of residuals (see section 7.6.1.1) as new variables in the data editor. These residual variables can then be examined to see how well the model fits the observed data. To save residuals click on in the main Logistic Regression dialog box (Figure 8.3). SPSS saves each of the selected variables into the data editor but they can be listed in the output viewer by using the Case Summaries command (see section 7.8.6) and selecting the residual variables of interest. The residuals dialog box in Figure 8.5 gives us several options and most of these are the same as those in multiple regression (refer to section 7.7.5). Two residuals that are unique to logistic regression are the predicted probabilities and the predicted group memberships. The predicted probabilities are the probabilities of Y occurring given the values of each predictor for a given participant. As such, they are derived from equation (8.4) for a given case. The predicted group membership is self-explanatory in that it predicts to which of the two outcome categories a participant is most likely to belong based on the model. The group memberships are based on the predicted probabilities and I will explain these values in more detail when we consider how to interpret the residuals. It is worth selecting all of the available options, or as a bare minimum select the same options as in Figure 8.5.
To reiterate a point from the previous chapter, running a regression without checking how well the model fits the data is like buying a new pair of trousers without trying them on – they might look fine on the hanger but get them home and you find you’re Johnny-tight-pants. The trousers do their job (they cover your legs and keep you warm) but they have no real-life value (because they cut off the blood circulation to your legs, which then have to be amputated). Likewise, regression does its job regardless of the data – it will create a model – but the real-life value of the model may be limited (see section 7.6).
8.6. Interpreting logistic regression
8.6.1. The initial model
SPSS Output 8.1 tells us two things. First it tells us how we coded our outcome variable, and this table merely reminds us that 0 = not cured, and 1 = cured. [3] The second is that it tell us how SPSS has coded the categorical predictors. The parameter codings are also given for the categorical predictor variable (Intervention). Indicator coding was chosen with two categories, and so the coding is the same as the values in the data editor (0 = no treatment, 1 = treatment). If deviation coding had been chosen then the coding would have been -1 (Treatment) and 1 (No Treatment). With a simple contrast the codings would have been -0.5 (Intervention = No Treatment) and 0.5 (Intervention = Treatment) if [FIRST] was selected as the reference category or vice versa if [LAST] was selected as the reference category. The parameter codings are important for calculating the probability of the outcome variable (P(Y)), but we will come to that later.
For this first analysis we requested a forward stepwise method [4] and so the initial model is derived using only the constant in the regression equation. SPSS Output 8.2 tells us about the model when only the constant is included (i.e. all predictor variables are omitted). The table labelled Iteration History tells us that the log-likelihood of this baseline model (see section 8.3.1) is 154.08. This represents the fit of the most basic model to the data. When including only the constant, the computer bases the model on assigning every participant to a single category of the outcome variable. In this example, SPSS can decide either to predict that the patient was cured, or that every patient was not cured. It could make this decision arbitrarily, but because it is crucial to try to maximize how well the model predicts the observed data, SPSS will predict that every patient belongs to the category in which most observed cases fell. In this example there were 65 patients who were cured, and only 48 who were not cured. Therefore, if SPSS predicts that every patient was cured then this prediction will be correct 65 times out of 113 (i.e. 58% approx.). However, if SPSS predicted that every patient was not cured, then this prediction would be correct only 48 times out of 113 (42% approx.). As such, of the two available options it is better to predict that all patients were cured because this results in a greater number of correct predictions. The output shows a contingency table for the model in this basic state. You can see that SPSS has predicted that all patients are cured, which results in 0% accuracy for the patients who were not cured, and 100% accuracy for those observed to be cured. Overall, the model correctly classifies 57.5% of patients.
SPSS Output 8.3 summarizes the model (Variables in the Equation), and at this stage this entails quoting the value of the constant (b0), which is equal to 0.30. The table labelled Variables not in the Equation tells us that the residual chi-square statistic is 9.83 which is significant at p < .05 (it labels this statistic Overall Statistics). This statistic tells us that the coefficients for the variables not in the model are significantly different from zero – in other words, that the addition of one or more of these variables to the model will significantly affect its predictive power. If the probability for the residual chi-square had been greater than .05 it would have meant that forcing all of the variables excluded from the model into the model would not have made a significant contribution to its predictive power.
The remainder of this table lists each of the predictors in turn with a value of Roa’s efficient score statistic for each one (column labelled Score). In large samples when the null hypothesis is true, the score statistic is identical to the Wald statistic and the likelihood ratio statistic. It is used at this stage of the analysis because it is computationally less intensive than the Wald statistic and so can still be calculated in situations when the Wald statistic would prove prohibitive. Like any test statistic, Roa’s score statistic has a specific distribution from which statistical significance can be obtained. In this example, Intervention and the Intervention × Duration interaction both have significant score statistics at p < .01 and could potentially make a contribution to the model, but Duration alone does not look likely to be a good predictor because its score statistic is non-significant p > .05. As mentioned in section 8.5.2, the stepwise calculations are relative and so the variable that will be selected for inclusion is the one with the highest value for the score statistic that has a significance below .05. In this example, that variable will be Intervention because its score statistic (9.77) is the biggest.
[3] These values are the same as the data editor so this table might seem pointless; however, had we used codes other than 0 and 1 (e.g. 1 = not cured, 2 = cured) then SPSS changes these to zeros and ones and this table informs you of which category is represented by 0 and which by 1. This is important when it comes to interpretation.
[4] Actually, this is a really bad idea when you have an interaction term because to look at an interaction you need to include the main effects of the variables in the interaction term. I chose this method only to illustrate how stepwise methods work.
8.6.2. Step 1: intervention
As I predicted in the previous section, whether or not an intervention was given to the patient (Intervention) is added to the model in the first step. As such, a patient is now classified as being cured or not based on whether they had an intervention or not (waiting list). This can be explained easily if we look at the crosstabulation for the variables Intervention and Cured. [5] The model will use whether a patient had an intervention or not to predict whether they were cured or not by applying the crosstabulation table shown in Table 8.1.
The model predicts that all of the patients who had an intervention were cured. There were 57 patients who had an intervention, so the model predicts that these 57 patients were cured; it is correct for 41 of these patients, but misclassifies 16 people as ‘cured’ who were not cured – see Table 8.1. In addition, this new model predicts that all of the 56 patients who received no treatment were not cured; for these patients the model is correct 32 times but misclassifies as ‘not cured’ 24 people who were.
SPSS Output 8.4 shows summary statistics about the new model (which we’ve already seen contains Intervention). The overall fit of the new model is assessed using the loglikelihood statistic (see section 8.3.1). In SPSS, rather than reporting the log-likelihood itself, the value is multiplied by -2 (and sometimes referred to as -2LL): this multiplication is done because -2LL has an approximately chi-square distribution and so it makes it possible to compare values against those that we might expect to get by chance alone. Remember that large values of the log-likelihood statistic indicate poorly fitting statistical models.
At this stage of the analysis the value of -2LL should be less than the value when only the constant was included in the model (because lower values of -2LL indicate that the model is predicting the outcome variable more accurately). When only the constant was included, -2LL = 154.08, but now Intervention has been included this value has been reduced to 144.16. This reduction tells us that the model is better at predicting whether someone was cured than it was before Intervention was added. The question of how much better the model predicts the outcome variable can be assessed using the model chi-square statistic, which measures the difference between the model as it currently stands and the model when only the constant was included. We saw in section 8.3.1 that we could assess the significance of the change in a model by taking the log-likelihood of the new model and subtracting the log-likelihood of the baseline model from it. The value of the model chisquare statistic works on this principle and is, therefore, equal to -2LL with Intervention included minus the value of -2LL when only the constant was in the model (154.08 – 144.16 = 9.92). This value has a chi-square distribution and so its statistical significance can be calculated easily. [6] In this example, the value is significant at a .05 level and so we can say that overall the model is predicting whether a patient is cured or not significantly better than it was with only the constant included. The model chi-square is an analogue of the F-test for the linear regression (see Chapter 7). In an ideal world we would like to see a non-significant overall -2LL (indicating that the amount of unexplained data is minimal) and a highly significant model chi-square statistic (indicating that the model including the predictors is significantly better than without those predictors). However, in reality it is possible for both statistics to be highly significant.
There is a second statistic called the step statistic that indicates the improvement in the predictive power of the model since the last stage. At this stage there has been only one step in the analysis and so the value of the improvement statistic is the same as the model chi-square. However, in more complex models in which there are three or four stages, this statistic gives a measure of the improvement of the predictive power of the model since the last step. Its value is equal to -2LL at the current step minus -2LL at the previous step. If the improvement statistic is significant then it indicates that the model now predicts the outcome significantly better than it did at the last step, and in a forward regression this can be taken as an indication of the contribution of a predictor to the predictive power of the model. Similarly, the block statistic provides the change in -2LL since the last block (for use in hierarchical or blockwise analyses).
SPSS Output 8.4 also tells us the values of Cox and Snell’s and Nagelkerke’s R², but we will discuss these a little later. There is also a classification table that indicates how well the model predicts group membership; because the model is using Intervention to predict the outcome variable, this classification table is the same as Table 8.1. The current model correctly classifies 32 patients who were not cured but misclassifies 16 others (it correctly classifies 66.7% of cases). The model also correctly classifies 41 patients who were cured but misclassifies 24 others (it correctly classifies 63.1% of cases). The overall accuracy of classification is, therefore, the weighted average of these two values (64.6%). So, when only the constant was included, the model correctly classified 57.5% of patients, but now, with the inclusion of Intervention as a predictor, this has risen to 64.6%.
The next part of the output (SPSS Output 8.5) is crucial because it tells us the estimates for the coefficients for the predictors included in the model. This section of the output gives us the coefficients and statistics for the variables that have been included in the model at this point (namely Intervention and the constant). The b-value is the same as the b-value in linear regression: they are the values that we need to replace in equation (8.4) to establish the probability that a case falls into a certain category. We saw in linear regression that the value of b represents the change in the outcome resulting from a unit change in the predictor variable. The interpretation of this coefficient in logistic regression is very similar in that it represents the change in the logit of the outcome variable associated with a one-unit change in the predictor variable. The logit of the outcome is simply the natural logarithm of the odds of Y occurring.
The crucial statistic is the Wald statistic [7] which has a chi-square distribution and tells us whether the b coefficient for that predictor is significantly different from zero. If the coefficient is significantly different from zero then we can assume that the predictor is making a significant contribution to the prediction of the outcome (Y). We came across the Wald statistic in section 8.3.3 and saw that it should be used cautiously because when the regression coefficient (b) is large, the standard error tends to become inflated, resulting in the Wald statistic being underestimated (see Menard, 1995). However, for these data it seems to indicate that having the intervention (or not) is a significant predictor of whether the patient is cured (note that the significance of the Wald statistic is less than .05).
In section 8.3.2 we saw that we could calculate an analogue of R using equation (8.7). For these data, the Wald statistic and its df can be read from SPSS Output 8.5 (9.45 and 1 respectively), and the original -2LL was 154.08. Therefore, R can be calculated as:
R = ± SQRT (9.45 − (2 × 1) / 154.08) (8.14)
= .22
In the same section we saw that Hosmer and Lemeshow’s measure (R²L) is calculated by dividing the model chi-square by the original -2LL. In this example the model chi-square after Intervention has been entered into the model is 9.93, and the original -2LL (before any variables were entered) was 154.08. So, R²L = 9.93/154.08 = .06, which is different to the value we would get by squaring the value of R given above (R² = .22² = 0.05). Earlier on in SPSS Output 8.4, SPSS gave us two other measures of R² that were described in section 8.3.2. The first is Cox and Snell’s measure, which SPSS reports as .084, and the second is Nagelkerke’s adjusted value, which SPSS reports as .113. As you can see, all of these values differ, but they can be used as effect size measures for the model.
SELF-TEST Using equations (8.9) and (8.10) calculate the values of Cox and Snell’s and Nagelkerke’s R² reported by SPSS. [Hint: These equations use the log-likelihood, whereas SPSS reports -2 × log-likelihood. LL(new) is, therefore, 144.16/−2 = -72.08, and LL(baseline) = 154.08/−2 = -77.04. The sample size, n, is 113.]
The final thing we need to look at is the odds ratio (Exp(B) in the SPSS output), which was described in section 8.3.4. To calculate the change in odds that results from a unit change in the predictor for this example, we must first calculate the odds of a patient being cured given that they didn’t have the intervention. We then calculate the odds of a patient being cured given that they did have the intervention. Finally, we calculate the proportionate change in these two odds.
To calculate the first set of odds, we need to use equation (8.12) to calculate the probability of a patient being cured given that they didn’t have the intervention. The parameter coding at the beginning of the output told us that patients who did not have the intervention were coded with a 0, so we can use this value in place of X. The value of b1has been estimated for us as 1.229 (see Variables in the Equation in SPSS Output 8.5), and the coefficient for the constant can be taken from the same table and is -0.288. We can calculate the odds as:
P(Cured) = 1 / 1 + e − (b0 + b1X1)
= 1 / 1 + e − [−0.288+(1.299×0)]
= 0.428
P(Not Cured) = 1 − P(Cured)
= 1 − 0.428
= 0.572
odds = 0.428 / 0.572
= 0.748
(8.15)
Now, we calculate the same thing after the predictor variable has changed by one unit. In this case, because the predictor variable is dichotomous, we need to calculate the odds of a patient being cured, given that they have had the intervention. So, the value of the intervention variable, X, is now 1 (rather than 0). The resulting calculations are as follows:
P(Cured) = 1 / 1 + e − (b0 + b1X1)
= 1 / 1 + e − [−0.288+(1.299×1)]
= 0.719
P(Not Cured) = 1 − P(Cured)
= 1 − 0.719
= 0.281
odds = 0.719 / 0.281
= 2.559
(8.16)
We now know the odds before and after a unit change in the predictor variable. It is now a simple matter to calculate the proportionate change in odds by dividing the odds after a unit change in the predictor by the odds before that change:
Δodds = odds after a unit change in the predictor / original odds
= 2.56 / 0.75
= 3.41
(8.17)
You should notice that the value of the proportionate change in odds is the same as the value that SPSS reports for Exp(B) (allowing for differences in rounding). We can interpret the odds ratio in terms of the change in odds. If the value is greater than 1 then it indicates that as the predictor increases, the odds of the outcome occurring increase. Conversely, a value less than 1 indicates that as the predictor increases, the odds of the outcome occurring decrease. In this example, we can say that the odds of a patient who is treated being cured are 3.41 times higher than those of a patient who is not treated.
In the options (see section 8.5.5), we requested a confidence interval for the odds ratio and it can also be found in the output. The way to interpret this confidence interval is the same as any other confidence interval (section 2.5.2): if we calculated confidence intervals for the value of the odds ratio in 100 different samples, then these intervals would encompass the actual value of the odds ratio in the population (rather than the sample) in 95 of those samples. In this case, we can be fairly confident that the population value of the odds ratio lies between 1.56 and 7.48. However, our sample could be one of the 5% that produces a confidence interval that ‘misses’ the population value.
The important thing about this confidence interval is that it doesn’t cross 1 (both values are greater than 1). This is important because values greater than 1 mean that as the predictor variable increases, so do the odds of (in this case) being cured. Values less than 1 mean the opposite: as the predictor variable increases, the odds of being cured decrease. The fact that both limits of our confidence interval are above 1 gives us confidence that the direction of the relationship that we have observed is true in the population (i.e. it’s likely that having an intervention compared to not increases the odds of being cured). If the lower limit had been below 1 then it would tell us that there is a chance that in the population the direction of the relationship is the opposite to what we have observed. This would mean that we could not trust that our intervention increases the odds of being cured.
The test statistics for Intervention if it were removed from the model are in SPSS Output 8.6. Now, remember that earlier on I said how the regression would place variables into the equation and then test whether they then met a removal criterion. Well, the Model if Term Removed part of the output tells us the effects of removal. The important thing to note is the significance value of the log-likelihood ratio (log LR). The log LR for this model is significant (p < .01) which tells us that removing Intervention from the model would have a significant effect on the predictive ability of the model – in other words, it would be a very bad idea to remove it!
Finally, we are told about the variables currently not in the model. First of all, the residual chi-square (labelled Overall Statistics in the output), which is non-significant, tells us that none of the remaining variables have coefficients significantly different from zero. Furthermore, each variable is listed with its score statistic and significance value, and for both variables their coefficients are not significantly different from zero (as can be seen from the significance values of .964 for Duration and .835 for the Duration×Intervention interaction). Therefore, no further variables will be added to the model.
SPSS Output 8.7 displays the classification plot that we requested in the options dialog box. This plot is a histogram of the predicted probabilities of a patient being cured. If the model perfectly fits the data, then this histogram should show all of the cases for which the event has occurred on the right-hand side, and all the cases for which the event hasn’t occurred on the left-hand side. In other words, all of the patients who were cured should appear on the right and all those who were not cured should appear on the left. In this example, the only significant predictor is dichotomous and so there are only two columns of cases on the plot. If the predictor is a continuous variable, the cases are spread out across many columns. As a rule of thumb, the more that the cases cluster at each end of the graph, the better; such a plot would show that when the outcome did actually occur (i.e. the patient was cured) the predicted probability of the event occurring is also high (i.e. close to 1). Likewise, at the other end of the plot it would show that when the event didn’t occur (i.e. the patient still had a problem) the predicted probability of the event occurring is also low (i.e. close to 0). This situation represents a model that is correctly predicting the observed outcome data. If, however, there are a lot of points clustered in the centre of the plot then it shows that for many cases the model is predicting a probability of .5 that the event will occur. In other words, for these cases there is little more than a 50:50 chance that the data are correctly predicted by the model – the model could predict these cases just as accurately by tossing a coin! In SPSS Output 8.7 it’s clear that cured cases are predicted relatively well by the model (the probabilities are not that close to .5), but for not cured cases the model is less good (the probability of classification is only slightly lower than .5 or chance!). Also, a good model will ensure that few cases are misclassified; in this example there are a few Ns (not cureds) appearing on the cured side, but more worryingly there are quite a few Cs (cureds) appearing on the N side.
[6] The degrees of freedom will be the number of parameters in the new model (the number of predictors plus 1, which in this case with one predictor means 2) minus the number of parameters in the baseline model (which is 1, the constant). So, in this case, df = 2 – 1 = 1.
[7] As we have seen, this is simply b divided by its standard error (1.229/.40 = 3.0725); however, SPSS actually quotes the Wald statistic squared. For these data 3.0725² = 9.44 as reported (within rounding error) in the table.
8.6.4. Interpreting residuals
Our conclusions so far are fine in themselves, but to be sure that the model is a good one, it is important to examine the residuals. In section 8.5.4 we saw how to get SPSS to save various residuals in the data editor. We can now interpret them.
We saw in the previous chapter that the main purpose of examining residuals in any regression is to (1) isolate points for which the model fits poorly, and (2) isolate points that exert an undue influence on the model. To assess the former we examine the residuals, especially the Studentized residual, standardized residual and deviance statistics. To assess the latter we use influence statistics such as Cook’s distance, DFBeta and leverage statistics. These statistics were explained in detail in section 7.6 and their interpretation in logistic regression is the same; therefore, Table 8.2 summarizes the main statistics that you should look at and what to look for, but for more detail consult the previous chapter.
If you request these residual statistics, SPSS saves them as new columns in the data editor. You can look at the values in the data editor, or produce a table using the ANALYZE, REPORTS, CASE SUMMARIES… dialog box.
The basic residual statistics for this example (Cook’s distance, leverage, standardized residuals and DFBeta values) are pretty good: note that all cases have DFBetas less than 1, and leverage statistics (LEV_1) are very close to the calculated expected value of 0.018. There are also no unusually high values of Cook’s distance (COO_1) which, all in all, means that there are no influential cases having an effect on the model. The standardized residuals all have values of less than ± 2 and so there seems to be very little here to concern us.
You should note that these residuals are slightly unusual because they are based on a single predictor that is categorical. This is why there isn’t a lot of variability in the values of the residuals. Also, if substantial outliers or influential cases had been isolated, you are not justified in eliminating these cases to make the model fit better. Instead these cases should be inspected closely to try to isolate a good reason why they were unusual. It might simply be an error in inputting data, or it could be that the case was one which had a special reason for being unusual: for example, there were other medical complications that might contribute to the constipation that were noted during the patient’s assessment. In such a case, you may have good reason to exclude the case and duly note the reasons why.
8.8.2. Testing for multicollinearity
In section 7.6.2.4 we saw how multicollinearity can affect the parameters of a regression model. Logistic regression is just as prone to the biasing effect of collinearity and it is essential to test for collinearity following a logistic regression analysis. Unfortunately, SPSS does not have an option for producing collinearity diagnostics in logistic regression (which can create the illusion that it is unnecessary to test for it!). However, you can obtain statistics such as the tolerance and VIF by simply running a linear regression analysis using the same outcome and predictors. For the penalty example in the previous section, access the Linear Regression dialog box by selecting ANALYZE, REGRESSION, LINEAR… . The completed dialog box is shown in Figure 8.8. It is unnecessary to specify lots of options (we are using this technique only to obtain tests of collinearity) but it is essential that you click on STATISTICS… and then select Collinearity diagnostics in the dialog box. Once you have selected COLLINEARITY DIAGNOSTICS, switch off all of the default options, click on CONTINUE to return to the Linear Regression dialog box, and then click on OK to run the analysis.
The results of the linear regression analysis are shown in SPSS Output 8.10. From the first table we can see that the tolerance values are 0.014 for Previous and Anxious and 0.575 for PSWQ. In Chapter 7 we saw various criteria for assessing collinearity. To recap, Menard (1995) suggests that a tolerance value less than 0.1 almost certainly indicates a serious collinearity problem. Myers (1990) also suggests that a VIF value greater than 10 is cause for concern and in these data the values are over 70 for both Anxious and Previous. It seems from these values that there is an issue of collinearity between the predictor variables. We can investigate this issue further by examining the collinearity diagnostics.
SPSS Output 8.10 also shows a table labelled Collinearity Diagnostics. In this table, we are given the eigenvalues of the scaled, uncentred cross-products matrix, the condition index and the variance proportions for each predictor. If any of the eigenvalues in this table are much larger than others then the uncentred cross-products matrix is said to be ill-conditioned, which means that the solutions of the regression parameters can be greatly affected by small changes in the predictors or outcome. In plain English, these values give us some idea as to how accurate our regression model is: if the eigenvalues are fairly similar then the derived model is likely to be unchanged by small changes in the measured variables. The condition indexes are another way of expressing these eigenvalues and represent the square root of the ratio of the largest eigenvalue to the eigenvalue of interest (so, for the dimension with the largest eigenvalue, the condition index will always be 1). For these data the final dimension has a condition index of 81.3, which is massive compared to the other dimensions. Although there are no hard and fast rules about how much larger a condition index needs to be to indicate collinearity problems, this case clearly shows that a problem exists.
The final step in analysing this table is to look at the variance proportions. The variance of each regression coefficient can be broken down across the eigenvalues and the variance proportions tell us the proportion of the variance of each predictor’s regression coefficient that is attributed to each eigenvalue. These proportions can be converted to percentages by multiplying them by 100 (to make them more easily understood). So, for example, for PSWQ 95% of the variance of the regression coefficient is associated with eigenvalue number 3, 4% is associated with eigenvalue number 2 and 1% is associated with eigenvalue number 1. In terms of collinearity, we are looking for predictors that have high proportions on the same small eigenvalue, because this would indicate that the variances of their regression coefficients are dependent. So we are interested mainly in the bottom few rows of the table (which represent small eigenvalues). In this example, 99% of the variance in the regression coefficients of both Anxiety and Previous is associated with eigenvalue number 4 (the smallest eigenvalue), which clearly indicates dependency between these variables.
The result of this analysis is pretty clear cut: there is collinearity between state anxiety and previous experience of taking penalties and this dependency results in the model becoming biased.
8.9.4. Interpreting the multinomial logistic regression output
The next part of the output (SPSS Output 8.12) relates to the fit of the model to the data. We know that the model is significantly better than no model, but is it a good fit of the data? The Pearson and deviance statistics test the same thing, which is whether the predicted values from the model differ significantly from the observed values. If these statistics are not significant then the predicted values are not significantly different from the observed values; in other words, the model is a good fit. Here we have contrasting results: the deviance statistic says that the model is a good fit of the data (p = .45, which is much higher than .05), but the Pearson test indicates the opposite, namely that predicted values are significantly different from the observed values (p < .001). Oh dear.
One answer is that differences between these statistics can be caused by overdispersion. This is a possibility that we need to look into. However, there are other reasons for this conflict: for example, the Pearson statistic can be very inflated by low expected frequencies (which could happen because we have so many empty cells as indicated by our warning). One thing that is certain is that conflicting deviance and Pearson chi-square statistics are not good news!
Let’s look into the possibility of overdispersion. We can compute the dispersion parameters from both statistics:
φPearson = χ²Pearson/df = 886.62/614 = 1.44
φDeviance = χ²Deviance/df = 617.48/614 = 1.01
Neither of these is particularly high, and the one based on the deviance statistic is close to the ideal value of 1. The value based on Pearson is greater than 1, but not close to 2, so again does not give us an enormous cause for concern that the data are overdispersed. [9]
The output also shows us the two other measures of R² that were described in section 8.3.2. The first is Cox and Snell’s measure, which SPSS reports as .24, and the second is Nagelkerke’s adjusted value, which SPSS reports as .28. As you can see, they are reasonably similar values and represent relatively decent-sized effects.
SPSS Output 8.13 shows the results of the likelihood ratio tests and these can be used to ascertain the significance of predictors to the model. The first thing to note is that no significance values are produced for covariates that are involved in higher-order interactions (this is why there are blank spaces in the Sig. column for the effects of Funny and Sex). This table tells us, though, that gender had a significant main effect on success rates of chat-up lines, χ²(2) = 18.54, p < .001, as did whether the chat-up lined showed evidence of being a good partner, χ²(2) = 6.32, p < .042. Most interesting are the interactions which showed that the humour in the chat-up line interacted with gender to predict success at getting a date, χ²(2) = 35.81, p < .001; also the sexual content of the chat-up line interacted with the gender of the person being chatted up in predicting their reaction, χ²(2) = 13.45, p < .001. These likelihood statistics can be seen as sorts of overall statistics that tell us which predictors significantly enable us to predict the outcome category, but they don’t really tell us specifically what the effect is. To see this we have to look at the individual parameter estimates.
Chapter 9
Comparing two means
JANE SUPERBRAIN 9.1 Are median splits the devil’s work?
Often in research papers you see that people have analysed their data using a ‘median split’. In our spider phobia example, this means that you measure scores on a spider phobia questionnaire and calculate the median. You then classify anyone with a score above the median as a ‘phobic’, and those below the median as ‘non-phobic’. In doing this you ‘dichotomize’ a continuous variable. This practice is quite common, but is it sensible?
MacCallum, Zhang, Preacher, and Rucker (2002) wrote a splendid paper pointing out various problems on turning a perfectly decent continuous variable into a categorical variable:
1. Imagine there are four people: Peter, Birgit, Jip and Kiki. We measure how scared of spiders they are as a percentage and get Jip (100%), Kiki (60%), Peter (40%) and Birgit (0%). If we split these four people at the median (50%) then we’re saying that Jip and Kiki are the same (they get a score of 1 = phobic) and Peter and Birgit are the same (they both get a score of 0 = not phobic). In reality, Kiki and Peter are the most similar of the four people, but they have been put in different groups. So, median splits change the original information quite dramatically (Peter and Kiki are originally very similar but become very different after the split, Jip and Kiki are relatively dissimilar originally but become identical after the split).
2. Effect sizes get smaller: if you correlate two continuous variables then the effect size will be larger than if you correlate the same variables after one of them has been dichotomized. Effect sizes also get smaller in ANOVA and regression.
3. There is an increased chance of finding spurious effects.
So, if your supervisor has just told you to do a median split, have a good think about whether it is the right thing to do (and read MacCallum et al.’s paper). One of the rare situations in which dichotomizing a continuous variable is justified, according to MacCallum et al., is when there is a clear theoretical rationale for distinct categories of people based on a meaningful break point (i.e. not the median); for example, phobic versus not phobic based on diagnosis by a trained clinician would be a legitimate dichotomization of anxiety.
9.7. The t-test as a general linear model
A lot of you might think it’s odd that I’ve chosen to represent the effect size for my t-tests using r, the correlation coefficient. In fact you might well be thinking ‘but correlations show relationships, not differences between means’. I used to think this too until I read a fantastic paper by Cohen (1968), which made me realize what I’d been missing; the complex, thorny, weed-infested and large Andy-eating tarantula-inhabited world of statistics suddenly turned into a beautiful meadow filled with tulips and little bleating lambs all jumping for joy at the wonder of life. Actually, I’m still a bumbling fool trying desperately to avoid having the blood sucked from my flaccid corpse by the tarantulas of statistics, but it was a good paper! What I’m about to say will either make no sense at all, or might help you to appreciate what I’ve said in most of the chapters so far: all statistical procedures are basically the same, they’re just more or less elaborate versions of the correlation coefficient!
In Chapter 7 we saw that the t-test was used to test whether the regression coefficient of a predictor was equal to zero. The experimental design for which the independent t-test is used can be conceptualized as a regression equation (after all, there is one independent variable (predictor) and one dependent variable (outcome)). If we want to predict our outcome, then we can use the general equation that I’ve mentioned at various points:
outcomei = (model) + errori
If we want to use a linear model, then we saw that this general equation becomes equation (7.2) in which the model is defined by the slope and intercept of a straight line. Equation (9.6) shows a very similar equation in which Ai is the dependent variable (outcome), b0 is the intercept, b1 is the weighting of the predictor and Gi is the independent variable (predictor). Now, I’ve also included the same equation but with some of the letters replaced with what they represent in the spider experiment (so, A = anxiety, G = group). When we run an experiment with two conditions, the independent variable has only two values (group 1 or group 2). There are several ways in which these groups can be coded (in the spider example we coded group 1 with the value 0 and group 2 with the value 1). This coding variable is known as a dummy variable and values of this variable represent groups of entities. We have come across this coding in section 7.11:
Ai = b0 + b1G1 + εi
Anxietyi = b0 + b1groupi + εi
(9.6)
Using the spider example, we know that the mean anxiety of the picture group was 40, and that the group variable is equal to 0 for this condition. Look at what happens when the group variable is equal to 0 (the picture condition): equation (9.6) becomes (if we ignore the residual term):
X̄Picture = b0 + (b1 × 0)
b0 = X̄Picture
b0 = 40
Therefore, b0 (the intercept) is equal to the mean of the picture group (i.e. it is the mean of the group coded as 0). Now let’s look at what happens when the group variable is equal to 1. This condition is the one in which a real spider was used, therefore the mean anxiety (X̄Real) of this condition was 47. Remembering that we have just found out that b0 is equal to the mean of the picture group (X̄Picture), equation (9.6) becomes:
X̄Real = b0 + (b1 × 1)
X̄Real = X̄Picture + b1
b1 = X̄Real − X̄Picture
= 47 − 40
= 7
b1, therefore, represents the difference between the group means. As such, we can represent a two-group experiment as a regression equation in which the coefficient of the independent variable (b1) is equal to the difference between group means, and the intercept (b0) is equal to the mean of the group coded as 0. In regression, the t-test is used to ascertain whether the regression coefficient (b1) is equal to 0, and when we carry out a t-test on grouped data we, therefore, test whether the difference between group means is equal to 0.
The resulting SPSS output should contain the regression summary table shown in SPSS Output 9.5. The first thing to notice is the value of the constant (b0): its value is 40, the same as the mean of the base category (the picture group). The second thing to notice is that the value of the regression coefficient b1 is 7, which is the difference between the two group means (47 – 40 = 7). Finally, the t-statistic, which tests whether b1 is significantly different from zero, is the same as for the independent t-test (see SPSS Output 9.4) and so is the significance value. [6]
[6] In fact, the value of the t-statistic is the same but has a positive sign rather than negative. You’ll remember from the discussion of the point–biserial correlation in section 6.5.5 that when you correlate a dichotomous variable the direction of the correlation coefficient depends entirely upon which cases are assigned to which groups. Therefore, the direction of the t-statistic here is similarly influenced by which group we select to be the base category (the category coded as 0).
9.8. What if my data are not normally distributed?
We’ve seen in this chapter that there are adjustments that can be made to the t-test when the assumption of homogeneity of variance is broken, but what about when you have non-normally distributed data? The first thing to note is that although a lot of early evidence suggested that t was accurate when distributions were skewed, the t-test can be biased when the assumption of normality is not met (Wilcox, 2005). Second, we need to remember that it’s the shape of the sampling distribution that matters, not the sample data. One option then is to use a big sample and rely on the central limit theorem (section 2.5.1) which says that the sampling distribution should be normal when samples are big. You could also try to correct the distribution using a transformation (but see Jane Superbrain Box 5.1). Another useful solution is to use one of a group of tests commonly referred to as non-parametric tests. These tests have fewer assumptions than their parametric counterparts and so are useful when your data violate the assumptions of parametric data described in Chapter 5. Some of these tests are described in Chapter 15. The non-parametric counterpart of the dependent t-test is called the Wilcoxon signed-rank Test (section 15.4), and the independent t-test has two non-parametric counterparts (both extremely similar) called the Wilcoxon rank-sum test and the Mann–Whitney test (section 15.3). I’d recommend reading these sections before moving on.
A final option is to use robust methods (see section 5.7.4). There are various robust ways to test differences between means that involve using trimmed means or a bootstrap. However, SPSS doesn’t do any of these directly. Should you wish to do these then plugin for SPSS. Look at the companion website for some demos of how to use the R plugin.
Chapter 9: Comparing two means
CRAMMING SAM’s Tips
• The independent t-test compares two means, when those means have come from different groups of entities; for example, if you have used different participants in each of two experimental conditions.
• Look at the column labelled Levene’s Test for Equality of Variance. If the Sig. value is less than .05 then the assumption of homogeneity of variance has been broken and you should look at the row in the table labelled Equal variances not assumed. If the Sig. value of Levene’s test is bigger than .05 then you should look at the row in the table labelled Equal variances assumed.
• Look at the column labelled Sig. If the value is less than .05 then the means of the two groups are significantly different.
• Look at the values of the means to tell you how the groups differ.
• SPSS provides only the two-tailed significance value; if you want the one-tailed significance just divide the value by 2.
• Report the t-statistic, the degrees of freedom and the significance value. Also report the means and their corresponding standard errors (or draw an error bar chart).
Chapter 10: Comparing several means, ANOVA (GLM 1)
10.2. The theory behind ANOVA
10.2.1. Inflated error rates
Before explaining how ANOVA works, it is worth mentioning why we don’t simply carry out several t-tests to compare all combinations of groups that have been tested. Imagine a situation in which there were three experimental conditions and we were interested in differences between these three groups. If we were to carry out t-tests on every pair of groups, then we would have to carry out three separate tests: one to compare groups 1 and 2, one to compare groups 1 and 3, and one to compare groups 2 and 3. If each of these t-tests uses a .05 level of significance then for each test the probability of falsely rejecting the null hypothesis (known as a Type I error) is only 5%. Therefore, the probability of no Type I errors is .95 (95%) for each test. If we assume that each test is independent (hence, we can multiply the probabilities) then the overall probability of no Type I errors is (.95)3= .95 × .95 × .95 = .857, because the probability of no Type I errors is .95 for each test and there are three tests. Given that the probability of no Type I errors is .857, then we can calculate the probability of making at least one Type I error by subtracting this number from 1 (remember that the maximum probability of any event occurring is 1). So, the probability of at least one Type I error is 1 – .857 = .143, or 14.3%. Therefore, across this group of tests, the probability of making a Type I error has increased from 5% to 14.3%, a value greater than the criterion accepted by social scientists. This error rate across statistical tests conducted on the same experimental data is known as the familywise or experimentwise error rate. An experiment with three conditions is a relatively simple design, and so the effect of carrying out several tests is not severe. If you imagine that we now increase the number of experimental conditions from three to five (which is only two more groups) then the number of t-tests that would need to done increases to 10. [2] The familywise error rate can be calculated using the general equation (10.1) below, in which n is the number of tests carried out on the data. With 10 tests carried out, the familywise error rate is .40 (1 – .9510 = .40), which means that there is a 40% chance of having made at least one Type I error. For this reason we use ANOVA rather than conducting lots of t-tests:
familywise error = 1 – (0.95)n (10.1)
10.2.2. Interpreting F
When we perform a t-test, we test the hypothesis that the two samples have the same mean. Similarly, ANOVA tells us whether three or more means are the same, so it tests the null hypothesis that all group means are equal. An ANOVA produces an F-statistic or F-ratio, which is similar to the t-statistic in that it compares the amount of systematic variance in the data to the amount of unsystematic variance. In other words, F is the ratio of the model to its error.
ANOVA is an omnibus test, which means that it tests for an overall experimental effect: so, there are things that ANOVA cannot tell us. Although ANOVA tells us whether the experimental manipulation was generally successful, it does not provide specific information about which groups were affected. Assuming an experiment was conducted with three different groups, the F-ratio tells us that the means of these three samples are not equal (i.e. that X̄1 = X̄2 = X̄3 is not true). However, there are several ways in which the means can differ. The first possibility is that all three sample means are significantly different (X̄1 ≠ X̄2 ≠ X̄3). A second possibility is that the means of group 1 and 2 are the same but group 3 has a significantly different mean from both of the other groups (X̄1 = X̄2 ≠ X̄3). Another possibility is that groups 2 and 3 have similar means but group 1 has a significantly different mean (X̄1 ≠ X̄2 = X̄3). Finally, groups 1 and 3 could have similar means but group 2 has a significantly different mean from both (X̄1 = X̄3 ≠ X̄2). So, in an experiment, the F-ratio tells us only that the experimental manipulation has had some effect, but it doesn’t tell us specifically what the effect was.
10.2.3. ANOVA as regression
I’ve hinted several times that all statistical tests boil down to variants on regression. In fact, ANOVA is just a special case of regression. This surprises many scientists because ANOVA and regression are usually used in different situations. The reason is largely historical in that two distinct branches of methodology developed in the social sciences: correlational research and experimental research. Researchers interested in controlled experiments adopted ANOVA as their statistic of choice whereas those looking for real-world relationships adopted multiple regression. As we all know, scientists are intelligent, mature and rational people and so neither group was tempted to slag off the other and claim that their own choice of methodology was far superior to the other (yeah right!). With the divide in methodologies came a chasm between the statistical methods adopted by the two opposing camps (Cronbach, 1957, documents this divide in a lovely article). This divide has lasted many decades to the extent that now students are generally taught regression and ANOVA in very different contexts and many textbooks teach ANOVA in an entirely different way to regression.
Although many considerably more intelligent people than me have attempted to redress the balance (notably the great Jacob Cohen, 1968), I am passionate about making my own small, feeble-minded attempt to enlighten you (and I set the ball rolling in sections 7.11 and 9.7). There are several good reasons why I think ANOVA should be taught within the context of regression. First, it provides a familiar context: I wasted many trees trying to explain regression, so why not use this base of knowledge to explain a new concept (it should make it easier to understand)? Second, the traditional method of teaching ANOVA (known as the variance-ratio method) is fine for simple designs, but becomes impossibly cumbersome in more complex situations (such as analysis of covariance). The regression model extends very logically to these more complex designs without anyone needing to get bogged down in mathematics. Finally, the variance-ratio method becomes extremely unmanageable in unusual circumstances such as when you have unequal sample sizes. [3] The regression method makes these situations considerably simpler. Although these reasons are good enough, it is also the case that SPSS has moved away from the variance-ratio method of ANOVA and progressed towards solely using the regression model (known as the general linear model, or GLM).
I have mentioned that ANOVA is a way of comparing the ratio of systematic variance to unsystematic variance in an experimental study. The ratio of these variances is known as the F-ratio. However, any of you who have read Chapter 7 should recognize the F-ratio (see section 7.2.3) as a way to assess how well a regression model can predict an outcome compared to the error within that model. If you haven’t read Chapter 7 (surely not!), have a look before you carry on (it should only take you a couple of weeks to read). How can the F-ratio be used to test differences between means and whether a regression model fits the data? The answer is that when we test differences between means we are fitting a regression model and using F to see how well it fits the data, but the regression model contains only categorical predictors (i.e. grouping variables). So, just as the t-test could be represented by the linear regression equation (see section 9.7), ANOVA can be represented by the multiple regression equation in which the number of predictors is one less than the number of categories of the independent variable.
Let’s take an example. There was a lot of controversy, when I wrote the first edition of this book, surrounding the drug Viagra. Admittedly there’s less controversy now, but the controversy has been replaced by an alarming number of spam emails on the subject (for which I’ll no doubt be grateful in 20 years’ time), so I’m going to stick with the example. Viagra is a sexual stimulant (used to treat impotence) that broke into the black market under the belief that it will make someone a better lover (oddly enough, there was a glut of journalists taking the stuff at the time in the name of ‘investigative journalism’ … hmmm!). Suppose we tested this belief by taking three groups of participants and administering one group with a placebo (such as a sugar pill), one group with a low dose of Viagra and one with a high dose. The dependent variable was an objective measure of libido (I will tell you only that it was measured over the course of a week – the rest I will leave to your own imagination). The data can be found in the file Viagra.sav (which is described in detail later in this chapter) and are in Table 10.1.
If we want to predict levels of libido from the different levels of Viagra then we can use the general equation that keeps popping up:
outcomei = (model) + errori
If we want to use a linear model, then we saw in section 9.7 that when there are only two groups we could replace the ‘model’ in this equation with a linear regression equation with one dummy variable to describe two experimental groups. This dummy variable was a categorical variable with two numeric codes (0 for one group and 1 for the other). With three groups, however, we can extend this idea and use a multiple regression model with two dummy variables. In fact, as a general rule we can extend the model to any number of groups and the number of dummy variables needed will be one less than the number of categories of the independent variable. In the two-group case, we assigned one category as a base category (remember that in section 9.7 we chose the picture condition to act as a base) and this category was coded with 0. When there are three categories we also need a base category and you should choose the condition to which you intend to compare the other groups. Usually this category will be the control group. In most well-designed social science experiments there will be a group of participants who act as a baseline for other categories. This baseline group should act as the reference or base category, although the group you choose will depend upon the particular hypotheses that you want to test. In unbalanced designs (in which the group sizes are unequal) it is important that the base category contains a fairly large number of cases to ensure that the estimates of the regression coefficients are reliable. In the Viagra example, we can take the placebo group as the base category because this group was a placebo control. We are interested in comparing both the high- and low-dose groups to the group which received no Viagra at all. If the placebo group is the base category then the two dummy variables that we have to create represent the other two conditions: so, we should have one dummy variable called High and the other one called Low). The resulting equation is described as:
Libidoi = b0 + b2Highi + b1Lowi + εi (10.2)
In equation (10.2), a person’s libido can be predicted from knowing their group code (i.e. the code for the High and Low dummy variables) and the intercept (b0) of the model. The dummy variables in equation (10.2) can be coded in several ways, but the simplest way is to use a similar technique to that of the t-test. The base category is always coded as 0. If a participant was given a high dose of Viagra then they are coded with a 1 for the High dummy variable and 0 for all other variables. If a participant was given a low dose of Viagra then they are coded with the value 1 for the Low dummy variable and coded with 0 for all other variables (this is the same type of scheme we used in section 7.11). Using this coding scheme we can express each group by combining the codes of the two dummy variables (see Table 10.2).
Placebo group: Let’s examine the model for the placebo group. In the placebo group both the High and Low dummy variables are coded as 0. Therefore, if we ignore the error term (εi), the regression equation becomes:
Libidoi = b0 + (b2×0) + (b1×0)
Libidoi = b0
X̄Placebo = b0
This is a situation in which the high- and low-dose groups have both been excluded (because they are coded with 0). We are looking at predicting the level of libido when both doses of Viagra are ignored, and so the predicted value will be the mean of the placebo group (because this group is the only one included in the model). Hence, the intercept of the regression model, b0, is always the mean of the base category (in this case the mean of the placebo group).
High-dose group: If we examine the high-dose group, the dummy variable High will be coded as 1 and the dummy variable Low will be coded as 0. If we replace the values of these codes into equation (10.2) the model becomes:
Libidoi = b0 + (b2×1) + (b1×0)
Libidoi = b0 + b2
We know already that b0 is the mean of the placebo group. If we are interested in only the high-dose group then the model should predict that the value of Libido for a given participant equals the mean of the high-dose group. Given this information, the equation becomes:
Libidoi = b0 + b2
X̄High = X̄Placebo + b2
b2 = X̄High − X̄Placebo
Hence, b2 represents the difference between the means of the high-dose group and the placebo group.
Low-dose group: Finally, if we look at the model when a low dose of Viagra has been taken, the dummy variable Low is coded as 1 (and hence High is coded as 0). Therefore, the regression equation becomes:
Libidoi = b0 +(b2×0) + (b1×1)
Libidoi = b0 + b1
We know that the intercept is equal to the mean of the base category and that for the low-dose group the predicted value should be the mean libido for a low dose. Therefore the model can be reduced down to:
Libidoi = b0 + b1
X̄Low = X̄Placebo + b1
b1 = X̄Low − X̄Placebo
Hence, b1 represents the difference between the means of the low-dose group and the placebo group. This form of dummy variable coding is the simplest form, but as we will see later, there are other ways in which variables can be coded to test specific hypotheses. These alternative coding schemes are known as contrasts (see section 10.2.11.2). The idea behind contrasts is that you code the dummy variables in such a way that the b-values represent differences between groups that you are interested in testing.
The resulting analysis is shown in SPSS Output 10.1. It might be a good idea to remind yourself of the group means from Table 10.1. The first thing to notice is that just as in the regression chapter, an ANOVA has been used to test the overall fit of the model. This test is significant, F(2, 12) = 5.12, p < .05. Given that our model represents the group differences, this ANOVA tells us that using group means to predict scores is significantly better than using the overall mean: in other words, the group means are significantly different.
In terms of the regression coefficients, bs, the constant is equal to the mean of the base category (the placebo group). The regression coefficient for the first dummy variable (b2) is equal to the difference between the means of the high-dose group and the placebo group (5.0 − 2.2 = 2.8). Finally, the regression coefficient for the second dummy variable (b1) is equal to the difference between the means of the low-dose group and the placebo group (3.2 − 2.2 = 1). This analysis demonstrates how the regression model represents the three-group situation. We can see from the significance values of the t-tests that the difference between the high-dose group and the placebo group (b2) is significant because p < .05. The difference between the low-dose and the placebo group is not, however, significant (p = .282).
A four-group experiment can be described by extending the three-group scenario. I mentioned earlier that you will always need one less dummy variable than the number of groups in the experiment: therefore, this model requires three dummy variables. As before, we need to specify one category that is a base category (a control group). This base category should have a code of 0 for all three dummy variables. The remaining three conditions will have a code of 1 for the dummy variable that described that condition and a code of 0 for the other two dummy variables. Table 10.3 illustrates how the coding scheme would work.
10.2.4. Logic of the F-ratio
In Chapter 7 we learnt a little about the F-ratio and its calculation. To recap, we learnt that the F-ratio is used to test the overall fit of a regression model to a set of observed data. In other words, it is the ratio of how good the model is compared to how bad it is (its error). I have just explained how ANOVA can be represented as a regression equation, and this should help you to understand what the F-ratio tells you about your data. Figure 10.2 shows the Viagra data in graphical form (including the group means, the overall mean and the difference between each case and the group mean). In this example, there were three groups; therefore, we want to test the hypothesis that the means of three groups are different (so, the null hypothesis is that the group means are the same). If the group means were all the same, then we would not expect the placebo group to differ from the low-dose group or the high-dose group, and we would not expect the low-dose group to differ from the high-dose group. Therefore, on the diagram, the three coloured lines would be in the same vertical position (the exact position would be the grand mean – the dashed line in the figure). We can see from the diagram that the group means are actually different because the coloured lines (the group means) are in different vertical positions. We have just found out that in the regression model, b2 represents the difference between the means of the placebo and the high-dose group, and b1 represents the difference in means between the low-dose and placebo groups. These two distances are represented in Figure 10.2 by the vertical arrows. If the null hypothesis is true and all the groups have the same means, then these b coefficients should be zero (because if the group means are equal then the difference between them will be zero).
The logic of ANOVA follows from what we understand about regression:
• The simplest model we can fit to a set of data is the grand mean (the mean of the outcome variable). This basic model represents ‘no effect’ or ‘no relationship between the predictor variable and the outcome’.
• We can fit a different model to the data collected that represents our hypotheses. If this model fits the data well then it must be better than using the grand mean. Sometimes we fit a linear model (the line of best fit) but in experimental research we often fit a model based on the means of different conditions.
• The intercept and one or more regression coefficients can describe the chosen model.
• The regression coefficients determine the shape of the model that we have fitted; therefore, the bigger the coefficients, the greater the deviation between the line and the grand mean.
• In correlational research, the regression coefficients represent the slope of the line, but in experimental research they represent the differences between group means.
• The bigger the differences between group means, the greater the difference between the model and the grand mean.
• If the differences between group means are large enough, then the resulting model will be a better fit of the data than the grand mean.
• If this is the case we can infer that our model (i.e. predicting scores from the group means) is better than not using a model (i.e. predicting scores from the grand mean). Put another way, our group means are significantly different.
Just like when we used ANOVA to test a regression model, we can compare the improvement in fit due to using the model (rather than the grand mean) to the error that still remains. Another way of saying this is that when the grand mean is used as a model, there will be a certain amount of variation between the data and the grand mean. When a model is fitted it will explain some of this variation but some will be left unexplained. The F-ratio is the ratio of the explained to the unexplained variation. Look back at section 7.2.3 to refresh you memory on these concepts before reading on. This may all sound quite complicated, but actually most of it boils down to variations on one simple equation (see Jane Superbrain Box 10.1).
JANE SUPERBRAIN 10.1 You might be surprised to know that ANOVA boils down to one equation (well, sort of)
At every stage of the ANOVA we’re assessing variation (or deviance) from a particular model (be that the most basic model, or the most sophisticated model). We saw back in section 2.4.1 that the extent to which a model deviates from the observed data can be expressed, in general, in the form of equation (10.3). So, in ANOVA, as in regression, we use equation (10.3) to calculate the fit of the most basic model, and then the fit of the best model (the line of best fit). If the best model is any good then it should fit the data significantly better than our basic model:
deviation = Σ(observed – model)² (10.3)
The interesting point is that all of the sums of squares in ANOVA are variations on this one basic equation. All that changes is what we use as the model, and what the corresponding observed data are. Look through the various sections on the sums of squares and compare the resulting equations to equation (10.3); hopefully, you can see that they are all basically variations on this general form of the equation!
10.2.10. Assumptions of ANOVA
The assumptions under which the F statistic is reliable are the same as for all parametric tests based on the normal distribution (see section 5.2). That is, the variances in each experimental condition need to be fairly similar, observations should be independent and the dependent variable should be measured on at least an interval scale. In terms of normality, what matters is that distributions within groups are normally distributed.
You often hear people say ‘ANOVA is a robust test’, which means that it doesn’t matter much if we break the assumptions of the test: the F will still be accurate. There is some truth to this statement, but it is also an oversimplification of the situation. For one thing, the term ANOVA covers many different situations and the performance of F has been investigated in only some of those situations. There are two issues to consider: (1) does the F control the Type I error rate or is it significant even when there are no differences between means; and (2) does the F have enough power (i.e. is it able to detect differences when they are there)? Let’s have a look at the evidence.
Looking at normality first, Glass et al. (1972) reviewed a lot of evidence that suggests that F controls the Type I error rate well under conditions of skew, kurtosis and non-normality. Skewed distributions seem to have little effect on the error rate and power for two-tailed tests (but can have serious consequences for one-tailed tests). However, some of this evidence has been questioned (see Jane Superbrain Box 5.1). In terms of kurtosis, leptokurtic distributions make the Type I error rate too low (too many null effects are significant) and consequently the power is too high; platykurtic distributions have the opposite effect. The effects of kurtosis seem unaffected by whether sample sizes are equal or not. One study that is worth mentioning in a bit of detail is by Lunney (1970) who investigated the use of ANOVA in about the most nonnormal situation you could imagine: when the dependent variable is binary (it could have values of only 0 or 1). The results showed that when the group sizes were equal, ANOVA was accurate when there were at least 20 degrees of freedom and the smallest response category contained at least 20% of all responses. If the smaller response category contained less than 20% of all responses then ANOVA performed accurately only when there were 40 or more degrees of freedom. The power of F also appears to be relatively unaffected by non-normality (Donaldson, 1968). This evidence suggests that when group sizes are equal the F-statistic can be quite robust to violations of normality.
However, when group sizes are not equal the accuracy of F is affected by skew, and non-normality also affects the power of F in quite unpredictable ways (Wilcox, 2005). One situation that Wilcox describes shows that when means are equal the error rate (which should be 5%) can be as high as 18%. If you make the differences between means bigger you should find that power increases, but actually he found that initially power decreased (although it increased when he made the group differences bigger still). As such F can be biased when normality is violated.
In terms of violations of the assumption of homogeneity of variance, ANOVA is fairly robust in terms of the error rate when sample sizes are equal. However, when sample sizes are unequal, ANOVA is not robust to violations of homogeneity of variance (this is why earlier on I said it’s worth trying to collect equal-sized samples of data across conditions!). When groups with larger sample sizes have larger variances than the groups with smaller sample sizes, the resulting F-ratio tends to be conservative. That is, it’s more likely to produce a non-significant result when a genuine difference does exist in the population. Conversely, when the groups with larger sample sizes have smaller variances than the groups with smaller samples sizes, the resulting F-ratio tends to be liberal. That is, it is more likely to produce a significant result when there is no difference between groups in the population (put another way, the Type I error rate is not controlled) – see Glass et al. (1972) for a review. When variances are proportional to the means then the power of F seems to be unaffected by the heterogeneity of variance and trying to stabilize variances does not substantially improve power (Budescu, 1982; Budescu & Appelbaum, 1981). Problems resulting from violations of homogeneity of variance assumption can be corrected (see Jane Superbrain Box 10.2).
Violations of the assumption of independence are very serious indeed. Scariano and Davenport (1987) showed that when this assumption is violated (i.e. observations across groups are correlated) then the Type I error rate is substantially inflated. For example, using the conventional .05 Type I error rate when observations are independent, if these observations are made to correlate moderately (say, with a Pearson coefficient of .5), when comparing three groups to 10 observations per group the actual Type I error rate is .74 (a substantial inflation!). Therefore, if observations are correlated you might think that you are working with the accepted .05 error rate (i.e. you’ll incorrectly find a significant result only 5% of the time) when in fact your error rate is closer to .75 (i.e. you’ll find a significant result on 75% of occasions when, in reality, there is no effect in the population)!
Chapter 11: Analysis of covariance, ANCOVA (GLM 2)
11.3.1. Independence of the covariate and treatment effect
I said in the previous section that one use of ANCOVA is to reduce within-group error variance by allowing the covariate to explain some of this error variance. However, for this to be true the covariate must be independent from the experimental effect.
Figure 11.2 shows three different scenarios. Part A shows a basic ANOVA and is similar to Figure 10.3; it shows that the experimental effect (in our example libido) can be partitioned into two parts that represent the experimental or treatment effect (in this case the administration of Viagra) and the error or unexplained variance (i.e. factors that affect libido that we haven’t measured). Part B shows the ideal scenario for ANCOVA in which the covariate shares its variance only with the bit of libido that is currently unexplained. In other words, it is completely independent from the treatment effect (it does not overlap with the effect of Viagra at all). This scenario is the only one in which ANCOVA is appropriate. Part C shows a situation in which people often use ANCOVA when they should not. In this situation the effect of the covariate overlaps with the experimental effect. In other words, the experimental effect is confounded with the effect of the covariate. In this situation, the covariate will reduce (statistically speaking) the experimental effect because it explains some of the variance that would otherwise be attributable to the experiment. When the covariate and the experimental effect (independent variable) are not independent, the treatment effect is obscured, spurious treatment effects can arise and at the very least the interpretation of the ANCOVA is seriously compromised (Wildt & Ahtola, 1978).
The problem of the covariate and treatment sharing variance is common and is ignored or misunderstood by many people (Miller & Chapman, 2001). Miller and Chapman, in a very readable review, cite many situations in which people misapply ANCOVA and I recommend reading this paper. To summarize the main issue, when treatment groups differ on the covariate then putting the covariate into the analysis will not ‘control for’ or ‘balance out’ those differences (Lord, 1967, 1969). This situation arises mostly when participants are not randomly assigned to experimental treatment conditions. For example, anxiety and depression are closely correlated (anxious people tend to be depressed) so if you wanted to compare an anxious group of people against a non-anxious group on some task, the chances are that the anxious group would also be more depressed than the non-anxious group. You might think that by adding depression as a covariate into the analysis you can look at the ‘pure’ effect of anxiety but you can’t. This would be the situation in part C of Figure 11.2; the effect of the covariate (depression) would contain some of the variance from the effect of anxiety. Statistically speaking all that we know is that anxiety and depression share variance; we cannot separate this shared variance into ‘anxiety variance’ and ‘depression variance’, it will always just be ‘shared’. Another common example is if you happen to find that your experimental groups differ in their ages. Placing age into the analysis as a covariate will not solve this problem – it is still confounded with the experimental manipulation. ANCOVA is not a magic solution to this problem.
This problem can be avoided by randomizing participants to experimental groups, or by matching experimental groups on the covariate (in our anxiety example, you could try to find participants for the low anxious group who score high on depression). We can check whether this problem is likely to be an issue by checking whether experimental groups differ on the covariate before we run the ANCOVA. To use our anxiety example again, we could test whether our high and low anxious groups differ on levels of depression (with a t-test or ANOVA). If the groups do not significantly differ then we can use depression as a covariate.
Chapter 13: Repeated-measures designs (GLM 4)
13.3. Theory of one-way repeated-measures ANOVA
In a repeated-measures ANOVA the effect of our experiment is shown up in the withinparticipant variance (rather than in the between-group variance). Remember that in independent ANOVA (section 10.2) the within-participant variance is our residual variance (SSR); it is the variance created by individual differences in performance. This variance is not contaminated by the experimental effect, because whatever manipulation we’ve carried out has been done on different people. However, when we carry out our experimental manipulation on the same people then the within-participant variance will be made up of two things: the effect of our manipulation and, as before, individual differences in performance. So, some of the within-subjects variation comes from the effects of our experimental manipulation: we did different things in each experimental condition to the participants, and so variation in an individual’s scores will partly be due to these manipulations. For example, if everyone scores higher in one condition than another, it’s reasonable to assume that this happened not by chance, but because we did something different to the participants in one of the conditions compared to any other one. Because we did the same thing to everyone within a particular condition, any variation that cannot be explained by the manipulation we’ve carried out must be due to random factors outside our control, unrelated to our experimental manipulations (we could call this ‘error’). As in independent ANOVA, we use an F-ratio that compares the size of the variation due to our experimental manipulations to the size of the variation due to random factors, the only difference being how we calculate these variances. If the variance due to our manipulations is big relative to the variation due to random factors, we get a big value of F, and we can conclude that the observed results are unlikely to have occurred if there was no effect in the population.
Chapter 15: Non-parametric tests
15.2. When to use non-parametric tests
We’ve seen in the last few chapters how we can use various techniques to look for differences between means. However, all of these tests rely on parametric assumptions (see Chapter 5). Data are often unfriendly and don’t always turn up in nice normally distributed packages! Just to add insult to injury, it’s not always possible to correct for problems with the distribution of a data set – so, what do we do in these cases? The answer is that we have to use special kinds of statistical procedures known as non-parametric tests. Non-parametric tests are sometimes known as assumption-free tests because they make fewer assumptions about the type of data on which they can be used.[2] Most of these tests work on the principle of ranking the data: that is, finding the lowest score and giving it a rank of 1, then finding the next highest score and giving it a rank of 2, and so on. This process results in high scores being represented by large ranks, and low scores being represented by small ranks. The analysis is then carried out on the ranks rather than the actual data. This process is an ingenious way around the problem of using data that break the parametric assumptions. Some people believe that non-parametric tests have less power than their parametric counterparts, but as we will see in Jane Superbrain Box 15.1 below this is not always true. In this chapter we’ll look at four of the most common non-parametric procedures: the Mann–Whitney test, the Wilcoxon signed-rank test, Friedman’s test and the Kruskal–Wallis test. For each of these we’ll discover how to carry out the analysis on SPSS and how to interpret and report the results.
[2] Non-parametric tests sometimes get referred to as distribution-free tests, with an explanation that they make no assumptions about the distribution of the data. Technically, this isn’t true: they do make distributional assumptions (e.g. the ones in this chapter all assume a continuous distribution), but they are less restrictive ones than their parametric counterparts.
15.3. Comparing two independent conditions: the Wilcoxon rank-sum test and Mann-Whitney test
When you want to test differences between two conditions and different participants have been used in each condition then you have two choices: the Mann–Whitney test (Mann & Whitney, 1947) and the Wilcoxon rank-sum test (Wilcoxon, 1945; Figure 15.2). These tests are the non-parametric equivalent of the independent t-test. In fact both tests are equivalent, and there’s another, more famous, Wilcoxon test, so it gets extremely confusing for most of us. For example, a neurologist might collect data to investigate the depressant effects of certain recreational drugs. She tested 20 clubbers in all: 10 were given an ecstasy tablet to take on a Saturday night and 10 were allowed to drink only alcohol. Levels of depression were measured using the Beck Depression Inventory (BDI) the day after and midweek. The data are in Table 15.1.
15.3.1. Theory
The logic behind the Wilcoxon rank-sum and Mann–Whitney tests is incredibly elegant. First, let’s imagine a scenario in which there is no difference in depression levels between ecstasy and alcohol users. If we were to rank the data ignoring the group to which a person belonged from lowest to highest (i.e. give the lowest score a rank of 1 and the next lowest a rank of 2 etc.), then what should you find? Well, if there’s no difference between the groups then you expect to find a similar number of high and low ranks in each group; specifically, if you added up the ranks, then you’d expect the summed total of ranks in each group to be about the same. Now think about what would happen if there was a difference between the groups. Let’s imagine that the ecstasy group is more depressed than the alcohol group. If you rank the scores as before, then you would expect the higher ranks to be in the ecstasy group and the lower ranks to be in the alcohol group. Again, if we summed the ranks in each group, we’d expect the sum of ranks to be higher in the ecstasy group than in the alcohol group.
The Mann–Whitney and Wilcoxon rank-sum tests both work on this principle. In fact, when the groups have unequal numbers of participants in them then the test statistic (Ws) for the Wilcoxon rank-sum test is simply the sum of ranks in the group that contains the fewer people; when the group sizes are equal it’s the value of the smaller summed rank. Let’s have a look at how ranking works in practice.
Figure 15.3 shows the ranking process for both the Wednesday and Sunday data. To begin with, let’s use our data for Wednesday, because it’s more straightforward. First, just arrange the scores in ascending order, attach a label to remind you which group they came from (I’ve used A for alcohol and E for ecstasy), then starting at the lowest score assign potential ranks starting with 1 and going up to the number of scores you have. The reason why I’ve called these potential ranks is because sometimes the same score occurs more than once in a data set (e.g. in these data a score of 6 occurs twice, and a score of 35 occurs three times). These are called tied ranks and these values need to be given the same rank, so all we do is assign a rank that is the average of the potential ranks for those scores. So, with our two scores of 6, because they would’ve been ranked as 3 and 4, we take an average of these values (3.5) and use this value as a rank for both occurrences of the score! Likewise, with the three scores of 35, we have potential ranks of 16, 17 and 18; we actually use the average of these three ranks, (16+17+18)/3 = 17. When we’ve ranked the data, we add up all of the ranks for the two groups. So, add the ranks for the scores that came from the alcohol group (you should find the sum is 59) and then add the ranks for the scores that came from the ecstasy group (this value should be 151). We take the lowest of these sums to be our test statistic, therefore the test statistic for the Wednesday data is Ws = 59.
You should find that when you’ve ranked the data, and added the ranks for the two groups, the sum of ranks for the alcohol group is 90.5 and for the ecstasy group it is 119.5. The lowest of these sums is our test statistic; therefore the test statistic for the Sunday data is Ws = 90.5.
15.5.1. Theory of the Kruskal–Wallis test
The theory for the Kruskal–Wallis test is very similar to that of the Mann–Whitney (and Wilcoxon rank-sum) test, so before reading on look back at section 15.3.1. Like the Mann–Whitney test, the Kruskal–Wallis test is based on ranked data. So, to begin with, you simply order the scores from lowest to highest, ignoring the group to which the score belongs, and then assign the lowest score a rank of 1, the next highest a rank of 2 and so on (see section 15.3.1 for more detail). When you’ve ranked the data you collect the scores back into their groups and simply add up the ranks for each group. The sum of ranks for each group is denoted by Ri (where i is used to denote the particular group).
Table 15.3 shows the raw data for this example along with the ranks.
15.5.2. Inputting data and provisional analysis
When the data are collected using different participants in each group, we input the data using a coding variable. So, the data editor will have two columns of data. The first column is a coding variable (called something like Soya) which, in this case, will have four codes (for convenience I suggest 1 = no soya, 2 = one soya meal per week, 3 = four soya meals per week and 4 = seven soya meals per week). The second column will have values for the dependent variable (sperm count) measured at the end of the year (call this variable Sperm). When you enter the data into SPSS remember to tell the computer which group is represented by which code (see section 3.4.2.3). The data can be found in the file Soya.sav.
First, we would run some exploratory analyses on the data and because we’re going to be looking for group differences we need to run these exploratory analyses for each group. If you do these analyses you should find the same tables shown in SPSS Output 15.7. The first table shows that the Kolmogorov–Smirnov test (see section 5.5) was not significant for the control group (D(20) = .181, p > .05) but the Shapiro–Wilk test is significant and this test is actually more accurate (though less widely reported) than the Kolmogorov–Smirnov test (see Chapter 5). Data for the group that ate one soya meal per week were significantly different from normal (D(20) = .207, p < .05), as were the data for those that ate four (D(20) = .267, p < .01) and seven (D(20) = .204, p < .05). The second table shows the results of Levene’s test (section 5.6.1). The assumption of homogeneity of variance has been violated, F(3, 76) = 5.12, p < .01, and this is shown by the fact that the significance of Levene’s test is less than .05. As such, these data are not normally distributed, and the groups have heterogeneous variances!
Chapter 17: Exploratory factor analysis
17.3.3.2. Other methods
To overcome the problems associated with the regression technique, two adjustments have been proposed: the Bartlett method and the Anderson–Rubin method. SPSS can produce factor scores based on any of these methods. The Bartlett method produces scores that are unbiased and that correlate only with their own factor. The mean and standard deviation of the scores is the same as for the regression method. However, factor scores can still correlate with each other. The Anderson–Rubin method is a modification of the Bartlett method that produces factor scores that are uncorrelated and standardized (they have a mean of 0 and a standard deviation of 1). Tabachnick and Fidell (2007) conclude that the Anderson–Rubin method is best when uncorrelated scores are required but that the regression method is preferred in other circumstances simply because it is most easily understood. Although it isn’t important that you understand the maths behind any of the methods, it is important that you understand what the factor scores represent: namely, a composite score for each individual on a particular factor.
17.4.2. Communality
Before continuing it is important that you understand some basic things about the variance within an R-matrix. It is possible to calculate the variability in scores (the variance) for any given measure (or variable). You should be familiar with the idea of variance by now and comfortable with how it can be calculated (if not see Chapter 2). The total variance for a particular variable will have two components: some of it will be shared with other variables or measures (common variance) and some of it will be specific to that measure (unique variance). We tend to use the term unique variance to refer to variance that can be reliably attributed to only one measure. However, there is also variance that is specific to one measure but not reliably so; this variance is called error or random variance. The proportion of common variance present in a variable is known as the communality. As such, a variable that has no specific variance (or random variance) would have a communality of 1; a variable that shares none of its variance with any other variable would have a communality of 0.
In factor analysis we are interested in finding common underlying dimensions within the data and so we are primarily interested only in the common variance. Therefore, when we run a factor analysis it is fundamental that we know how much of the variance present in our data is common variance. This presents us with a logical impasse: to do the factor analysis we need to know the proportion of common variance present in the data, yet the only way to find out the extent of the common variance is by carrying out a factor analysis! There are two ways to approach this problem. The first is to assume that all of the variance is common variance. As such, we assume that the communality of every variable is 1. By making this assumption we merely transpose our original data into constituent linear components (known as principal component analysis). The second approach is to estimate the amount of common variance by estimating communality values for each variable. There are various methods of estimating communalities but the most widely used (including alpha factoring) is to use the squared multiple correlation (SMC) of each variable with all others. So, for the popularity data, imagine you ran a multiple regression using one measure (Selfish) as the outcome and the other five measures as predictors: the resulting multiple R² (see section 7.5.2) would be used as an estimate of the communality for the variable Selfish. This second approach is used in factor analysis. These estimates allow the factor analysis to be done. Once the underlying factors have been extracted, new communalities can be calculated that represent the multiple correlation between each variable and the factors extracted. Therefore, the communality is a measure of the proportion of variance explained by the extracted factors.
17.4.3. Factor analysis vs. principal component analysis
I have just explained that there are two approaches to locating underlying dimensions of a data set: factor analysis and principal component analysis. These techniques differ in the communality estimates that are used. Simplistically, though, factor analysis derives a mathematical model from which factors are estimated, whereas principal component analysis merely decomposes the original data into a set of linear variates (see Dunteman, 1989: Chapter 8, for more detail on the differences between the procedures). As such, only factor analysis can estimate the underlying factors and it relies on various assumptions for these estimates to be accurate. Principal component analysis is concerned only with establishing which linear components exist within the data and how a particular variable might contribute to that component. In terms of theory, this chapter is dedicated to principal component analysis rather than factor analysis. The reasons are that principal component analysis is a psychometrically sound procedure, it is conceptually less complex than factor analysis, and it bears numerous similarities to discriminant analysis (described in the previous chapter).
However, we should consider whether the techniques provide different solutions to the same problem. Based on an extensive literature review, Guadagnoli and Velicer (1988) concluded that the solutions generated from principal component analysis differ little from those derived from factor analytic techniques. In reality, there are some circumstances for which this statement is untrue. Stevens (2002) summarizes the evidence and concludes that with 30 or more variables and communalities greater than 0.7 for all variables, different solutions are unlikely; however, with fewer than 20 variables and any low communalities (< 0.4) differences can occur. The flip-side of this argument is eloquently described by Cliff (1987) who observed that proponents of factor analysis ‘insist that components analysis is at best a common factor analysis with some error added and at worst an unrecognizable hodgepodge of things from which nothing can be determined’ (p. 349). Indeed, feeling is strong on this issue with some arguing that when principal component analysis is used it should not be described as a factor analysis and that you should not impute substantive meaning to the resulting components. However, to non-statisticians the difference between a principal component and a factor may be difficult to conceptualize (they are both linear models), and the differences arise largely from the calculation.
17.4.4. Theory behind principal component analysis
Principal component analysis works in a very similar way to MANOVA and discriminant function analysis (see previous chapter). Although it isn’t necessary to understand the mathematical principles in any detail, readers of the previous chapter may benefit from some comparisons between the two techniques. For those who haven’t read that chapter, I suggest you flick through it before moving ahead!
In MANOVA, various sum of squares and cross-product matrices were calculated that contained information about the relationships between dependent variables. I mentioned before that these SSCP matrices could be easily converted to variance–covariance matrices, which represent the same information but in averaged form (i.e. taking account of the number of observations). I also said that by dividing each element by the relevant standard deviation the variance–covariance matrices becomes standardized. The result is a correlation matrix. In principal component analysis we usually deal with correlation matrices (although it is possible to analyse a variance–covariance matrix too) and the point to note is that this matrix pretty much represents the same information as an SSCP matrix in MANOVA. The difference is just that the correlation matrix is an averaged version of the SSCP that has been standardized.
In MANOVA, we used several SSCP matrices that represented different components of experimental variation (the model variation and the residual variation). In principal component analysis the covariance (or correlation) matrix cannot be broken down in this way (because all data come from the same group of participants). In MANOVA, we ended up looking at the variates or components of the SSCP matrix that represented the ratio of the model variance to the error variance. These variates were linear dimensions that separated the groups tested, and we saw that the dependent variables mapped onto these underlying components. In short, we looked at whether the groups could be separated by some linear combination of the dependent variables. These variates were found by calculating the eigenvectors of the SSCP. The number of variates obtained was the smaller of p (the number of dependent variables) or k – 1 (where k is the number of groups). In component analysis we do something similar (I’m simplifying things a little, but it will give you the basic idea). That is, we take a correlation matrix and calculate the variates. There are no groups of observations, and so the number of variates calculated will always equal the number of variables measured (p). The variates are described, as for MANOVA, by the eigenvectors associated with the correlation matrix. The elements of the eigenvectors are the weights of each variable on the variate (see equation (16.5)). These values are the factor loadings described earlier. The largest eigenvalue associated with each of the eigenvectors provides a single indicator of the substantive importance of each variate (or component). The basic idea is that we retain factors with relatively large eigenvalues and ignore those with relatively small eigenvalues.
In summary, component analysis works in a similar way to MANOVA. We begin with a matrix representing the relationships between variables. The linear components (also called variates, or factors) of that matrix are then calculated by determining the eigenvalues of the matrix. These eigenvalues are used to calculate eigenvectors, the elements of which provide the loading of a particular variable on a particular factor (i.e. they are the b-values in equation (17.1)). The eigenvalue is also a measure of the substantive importance of the eigenvector with which it is associated.
17.4.5. Factor extraction: eigenvalues and the scree plot
Not all factors are retained in an analysis, and there is debate over the criterion used to decide whether a factor is statistically important. I mentioned above that eigenvalues associated with a variate indicate the substantive importance of that factor. Therefore, it seems logical that we should retain only factors with large eigenvalues. How do we decide whether or not an eigenvalue is large enough to represent a meaningful factor? Well, one technique advocated by Cattell (1966b) is to plot a graph of each eigenvalue (Y-axis) against the factor with which it is associated (X-axis). This graph is known as a scree plot (because it looks like a rock face with a pile of debris, or scree, at the bottom). I mentioned earlier that it is possible to obtain as many factors as there are variables and that each has an associated eigenvalue. By graphing the eigenvalues, the relative importance of each factor becomes apparent. Typically there will be a few factors with quite high eigenvalues, and many factors with relatively low eigenvalues, so this graph has a very characteristic shape: there is a sharp descent in the curve followed by a tailing off (see Figure 17.3). Cattell (1966b) argued that the cut-off point for selecting factors should be at the point of inflexion of this curve. The point of inflexion is where the slope of the line changes dramatically: so, in Figure 17.3, imagine drawing a straight line that summarizes the vertical part of the plot and another that summarizes the horizontal part (the red dashed lines); then the point of inflexion is the data point at which these two lines meet. In both examples in Figure 17.3 the point of inflexion occurs at the third data point (factor); therefore, we would extract two factors. Thus, you retain (or extract) only factors to the left of the point of inflexion (and do not include the factor at the point of inflexion itself).[4] With a sample of more than 200 participants, the scree plot provides a fairly reliable criterion for factor selection (Stevens, 2002).
Although scree plots are very useful, factor selection should not be based on this criterion alone. Kaiser (1960) recommended retaining all factors with eigenvalues greater than 1. This criterion is based on the idea that the eigenvalues represent the amount of variation explained by a factor and that an eigenvalue of 1 represents a substantial amount of variation. Jolliffe (1972, 1986) reports that Kaiser’s criterion is too strict and suggests the third option of retaining all factors with eigenvalues more than 0.7. The difference between how many factors are retained using Kaiser’s methods compared to Jolliffe’s can be dramatic.
You might well wonder how the methods compare. Generally speaking, Kaiser’s criterion overestimates the number of factors to retain (see Jane Superbrain Box 17.2) but there is some evidence that it is accurate when the number of variables is less than 30 and the resulting communalities (after extraction) are all greater than 0.7. Kaiser’s criterion can also be accurate when the sample size exceeds 250 and the average communality is greater than or equal to 0.6. In any other circumstances you are best advised to use a scree plot provided the sample size is greater than 200 (see Stevens, 2002, for more detail). By default, SPSS uses Kaiser’s criterion to extract factors. Therefore, if you use the scree plot to determine how many factors are retained you may have to rerun the analysis specifying that SPSS extracts the number of factors you require.
However, as is often the case in statistics, the three criteria often provide different solutions! In these situations the communalities of the factors need to be considered. In principal component analysis we begin with communalities of 1 with all factors retained (because we assume that all variance is common variance). At this stage all we have done is to find the linear variates that exist in the data – so we have just transformed the data without discarding any information. However, to discover what common variance really exists between variables we must decide which factors are meaningful and discard any that are too trivial to consider. Therefore, we discard some information. The factors we retain will not explain all of the variance in the data (because we have discarded some information) and so the communalities after extraction will always be less than 1. The factors retained do not map perfectly onto the original variables – they merely reflect the common variance present in the data. If the communalities represent a loss of information then they are important statistics. The closer the communalities are to 1, the better our factors are at explaining the original data. It is logical that the more factors retained, the greater the communalities will be (because less information is discarded); therefore, the communalities are good indices of whether too few factors have been retained. In fact, with generalized least-squares factor analysis and maximum-likelihood factor analysis you can get a statistical measure of the goodness of fit of the factor solution (see the next chapter for more on goodness-of-fit tests). This basically measures the proportion of variance that the factor solution explains (so, can be thought of as comparing communalities before and after extraction).
As a final word of advice, your decision on how many factors to extract will depend also on why you’re doing the analysis; for example, if you’re trying to overcome multicollinearity problems in regression, then it might be better to extract too many factors than too few.
[4] Actually if you read Cattell’s original paper he advised including the factor at the point of inflexion as well because it is ‘desirable to include at least one common error factor as a “garbage can”’. The idea is that the point of inflexion represents an error factor. However, in practice this garbage can factor is rarely retained; also Thurstone argued that it is better to retain too few rather than too many factors so most people do not to retain the factor at the point of inflexion.
17.4.6. Improving interpretation: factor rotation
Once factors have been extracted, it is possible to calculate to what degree variables load onto these factors (i.e. calculate the loading of the variable on each factor). Generally, you will find that most variables have high loadings on the most important factor and small loadings on all other factors. This characteristic makes interpretation difficult, and so a technique called factor rotation is used to discriminate between factors. If a factor is a classification axis along which variables can be plotted, then factor rotation effectively rotates these factor axes such that variables are loaded maximally to only one factor. Figure 17.4 demonstrates how this process works using an example in which there are only two factors. Imagine that a sociologist was interested in classifying university lecturers as a demographic group. She discovered that two underlying dimensions best describe this group: alcoholism and achievement (go to any academic conference and you’ll see that academics drink heavily!). The first factor, alcoholism, has a cluster of variables associated with it (green circles) and these could be measures such as the number of units drunk in a week, dependency and obsessive personality. The second factor, achievement, also has a cluster of variables associated with it (blue circles) and these could be measures relating to salary, job status and number of research publications. Initially, the full lines represent the factors, and by looking at the co-ordinates it should be clear that the blue circles have high loadings for factor 2 (they are a long way up this axis) and medium loadings on factor 1 (they are not very far up this axis). Conversely, the green circles have high loadings for factor 1 and medium loadings for factor 2. By rotating the axes (dashed lines), we ensure that both clusters of variables are intersected by the factor to which they relate most. So, after rotation, the loadings of the variables are maximized onto one factor (the factor that intersects the cluster) and minimized on the remaining factor(s). If an axis passes through a cluster of variables, then these variables will have a loading of approximately zero on the opposite axis. If this idea is confusing, then look at Figure 17.4 and think about the values of the co-ordinates before and after rotation (this is best achieved by turning the book when you look at the rotated axes).
There are two types of rotation that can be done. The first is orthogonal rotation, and the left-hand side of Figure 17.4 represents this method. In Chapter 10 we saw that the term orthogonal means unrelated, and in this context it means that we rotate factors while keeping them independent, or unrelated. Before rotation, all factors are independent (i.e. they do not correlate at all) and orthogonal rotation ensures that the factors remain uncorrelated. That is why in Figure 17.4 the axes are turned while remaining perpendicular. [5] The other form of rotation is oblique rotation. The difference with oblique rotation is that the factors are allowed to correlate (hence, the axes of the right-hand diagram of Figure 17.4 do not remain perpendicular).
The choice of rotation depends on whether there is a good theoretical reason to suppose that the factors should be related or independent (but see my later comments on this), and also how the variables cluster on the factors before rotation. On the first point, we might not expect alcoholism to be completely independent of achievement (after all, high achievement leads to high stress, which can lead to the drinks cabinet!). Therefore, on theoretical grounds, we might choose oblique rotation. On the second point, Figure 17.4 demonstrates how the positioning of clusters is important in determining how successful the rotation will be (note the position of the green circles). Specifically, if an orthogonal rotation was carried out on the right-hand diagram it would be considerably less successful in maximizing loadings than the oblique rotation that is displayed. One approach is to run the analysis using both types of rotation. Pedhazur and Schmelkin (1991) suggest that if the oblique rotation demonstrates a negligible correlation between the extracted factors then it is reasonable to use the orthogonally rotated solution. If the oblique rotation reveals a correlated factor structure, then the orthogonally rotated solution should be discarded. In any case, an oblique rotation should be used only if there are good reasons to suppose that the underlying factors could be related in theoretical terms.
The mathematics behind factor rotation is complex (especially oblique rotation). However, in oblique rotation, because each factor can be rotated by different amounts a factor transformation matrix, Λ, is needed. The factor transformation matrix is a square matrix and its size depends on how many factors are extracted from the data. If two factors are extracted then it will be a 2 × 2 matrix, but if four factors are extracted then it becomes a 4 × 4 matrix. The values in the factor transformation matrix consist of sines and cosines of the angle of axis rotation (θ). This matrix is multiplied by the matrix of unrotated factor loadings, A, to obtain a matrix of rotated factor loadings.
For the case of two factors the factor transformation matrix would be:
Therefore, you should think of this matrix as representing the angle through which the axes have been rotated, or the degree to which factors have been rotated. The angle of rotation necessary to optimize the factor solution is found in an iterative way (see SPSS Tip 8.1) and different methods can be used.
[5] This term means that the axes are at right angles to one another.
17.4.6.1. Choosing a method of factor rotation
SPSS has three methods of orthogonal rotation (varimax, quartimax and equamax) and two methods of oblique rotation (direct oblimin and promax). These methods differ in how they rotate the factors and, therefore, the resulting output depends on which method you select. Quartimax rotation attempts to maximize the spread of factor loadings for a variable across all factors. Therefore, interpreting variables becomes easier. However, this often results in lots of variables loading highly onto a single factor. Varimax is the opposite in that it attempts to maximize the dispersion of loadings within factors. Therefore, it tries to load a smaller number of variables highly onto each factor resulting in more interpretable clusters of factors. Equamax is a hybrid of the other two approaches and is reported to behave fairly erratically (see Tabachnick and Fidell, 2007). For a first analysis, you should probably select varimax because it is a good general approach that simplifies the interpretation of factors.
The case with oblique rotations is more complex because correlation between factors is permitted. In the case of direct oblimin, the degree to which factors are allowed to correlate is determined by the value of a constant called delta. The default value in SPSS is 0 and this ensures that high correlation between factors is not allowed (this is known as direct quartimin rotation). If you choose to set delta to greater than 0 (up to 0.8), then you can expect highly correlated factors; if you set delta to less than 0 (down to -0.8) you can expect less correlated factors. The default setting of 0 is sensible for most analyses and I don’t recommend changing it unless you know what you are doing (see Pedhazur and Schmelkin, 1991: 620). Promax is a faster procedure designed for very large data sets.
In theory, the exact choice of rotation will depend largely on whether or not you think that the underlying factors should be related. If you expect the factors to be independent then you should choose one of the orthogonal rotations (I recommend varimax). If, however, there are theoretical grounds for supposing that your factors might correlate, then direct oblimin should be selected. In practice, there are strong grounds to believe that orthogonal rotations are a complete nonsense for naturalistic data, and certainly for any data involving humans (can you think of any psychological construct that is not in any way correlated with some other psychological construct?) As such, some argue that orthogonal rotations should never be used.
17.4.6.2. Substantive importance of factor loadings
Once a factor structure has been found, it is important to decide which variables make up which factors. Earlier I said that the factor loadings were a gauge of the substantive importance of a given variable to a given factor. Therefore, it makes sense that we use these values to place variables with factors. It is possible to assess the statistical significance of a factor loading (after all, it is simply a correlation coefficient or regression coefficient); however, there are various reasons why this option is not as easy as it seems (see Stevens, 2002: 393). Typically, researchers take a loading of an absolute value of more than 0.3 to be important. However, the significance of a factor loading will depend on the sample size. Stevens (2002) produced a table of critical values against which loadings can be compared. To summarize, he recommends that for a sample size of 50 a loading of 0.722 can be considered significant, for 100 the loading should be greater than 0.512, for 200 it should be greater than 0.364, for 300 it should be greater than 0.298, for 600 it should be greater than 0.21, and for 1000 it should be greater than 0.162. These values are based on an alpha level of .01 (two-tailed), which allows for the fact that several loadings will need to be tested (see Stevens, 2002, for further detail). Therefore, in very large samples, small loadings can be considered statistically meaningful. SPSS does not provide significance tests of factor loadings but by applying Stevens’ guidelines you should gain some insight into the structure of variables and factors.
The significance of a loading gives little indication of the substantive importance of a variable to a factor. This value can be found by squaring the factor loading to give an estimate of the amount of variance in a factor accounted for by a variable (like R²). In this respect Stevens (2002) recommends interpreting only factor loadings with an absolute value greater than 0.4 (which explain around 16% of the variance in the variable).
17.5.1. Before you begin
17.5.1.1. Sample size
Correlation coefficients fluctuate from sample to sample, much more so in small samples than in large. Therefore, the reliability of factor analysis is also dependent on sample size. Much has been written about the necessary sample size for factor analysis resulting in many ‘rules of thumb’. The common rule is to suggest that a researcher has at least 10–15 participants per variable. Although I’ve heard this rule bandied about on numerous occasions its empirical basis is unclear (however, Nunnally, 1978, did recommend having 10 times as many participants as variables). Kass and Tinsley (1979) recommended having between 5 and 10 participants per variable up to a total of 300 (beyond which test parameters tend to be stable regardless of the participant to variable ratio). Indeed, Tabachnick and Fidell (2007) agree that ‘it is comforting to have at least 300 cases for factor analysis’ (p. 613) and Comrey and Lee (1992) class 300 as a good sample size, 100 as poor and 1000 as excellent.
Fortunately, recent years have seen empirical research done in the form of experiments using simulated data (so-called Monte Carlo studies). Arrindell and van der Ende (1985) used real-life data to investigate the effect of different participant to variable ratios. They concluded that changes in this ratio made little difference to the stability of factor solutions. Guadagnoli and Velicer (1988) found that the most important factors in determining reliable factor solutions was the absolute sample size and the absolute magnitude of factor loadings. In short, they argue that if a factor has four or more loadings greater than 0.6 then it is reliable regardless of sample size. Furthermore, factors with 10 or more loadings greater than 0.40 are reliable if the sample size is greater than 150. Finally, factors with a few low loadings should not be interpreted unless the sample size is 300 or more. MacCallum, Widaman, Zhang, and Hong (1999) have shown that the minimum sample size or sample to variable ratio depends on other aspects of the design of the study. In short, their study indicated that as communalities become lower the importance of sample size increases. With all communalities above 0.6, relatively small samples (less than 100) may be perfectly adequate. With communalities in the 0.5 range, samples between 100 and 200 can be good enough provided there are relatively few factors each with only a small number of indicator variables. In the worst scenario of low communalities (well below 0.5) and a larger number of underlying factors they recommend samples above 500.
What’s clear from this work is that a sample of 300 or more will probably provide a stable factor solution but that a wise researcher will measure enough variables to adequately measure all of the factors that theoretically they would expect to find.
Another alternative is to use the Kaiser–Meyer–Olkin measure of sampling adequacy (KMO) (Kaiser, 1970). The KMO can be calculated for individual and multiple variables and represents the ratio of the squared correlation between variables to the squared partial correlation between variables. The KMO statistic varies between 0 and 1. A value of 0 indicates that the sum of partial correlations is large relative to the sum of correlations, indicating diffusion in the pattern of correlations (hence, factor analysis is likely to be inappropriate). A value close to 1 indicates that patterns of correlations are relatively compact and so factor analysis should yield distinct and reliable factors. Kaiser (1974) recommends accepting values greater than 0.5 as barely acceptable (values below this should lead you to either collect more data or rethink which variables to include). Furthermore, values between 0.5 and 0.7 are mediocre, values between 0.7 and 0.8 are good, values between 0.8 and 0.9 are great and values above 0.9 are superb (Hutcheson & Sofroniou, 1999).
17.5.1.2. Correlations between variables
When I was an undergraduate my statistics lecturer always used to say ‘if you put garbage in, you get garbage out’. This saying applies particularly to factor analysis because SPSS will always find a factor solution to a set of variables. However, the solution is unlikely to have any real meaning if the variables analysed are not sensible. The first thing to do when conducting a factor analysis or principal component analysis is to look at the intercorrelation between variables. There are essentially two potential problems: (1) correlations that are not high enough; and (2) correlations that are too high. The correlations between variables can be checked using the correlate procedure (see Chapter 6) to create a correlation matrix of all variables. This matrix can also be created as part of the main factor analysis. In both cases the remedy is to remove variables from the analysis. We will look at each problem in turn.
If our test questions measure the same underlying dimension (or dimensions) then we would expect them to correlate with each other (because they are measuring the same thing). Even if questions measure different aspects of the same things (e.g. we could measure overall anxiety in terms of sub-components such as worry, intrusive thoughts and physiological arousal), there should still be high intercorrelations between the variables relating to these sub-traits. We can test for this problem first by visually scanning the correlation matrix and looking for correlations below about .3 (you could use the significance of correlations but, given the large sample sizes normally used with factor analysis, this approach isn’t helpful because even very small correlations will be significant in large samples). If any variables have lots of correlations below .3 then consider excluding them. It should be immediately clear that this approach is very subjective: I’ve used fuzzy terms such as ‘about .3’ and ‘lots of ’, but I have to because every data set is different. Analysing data really is a skill, not a recipe book!
If you want an objective test of whether correlations (overall) are too small then we can test for a very extreme scenario. If the variables in our correlation matrix did not correlate at all, then our correlation matrix would be an identity matrix (i.e. the off-diagonal components are zero – see section 16.4.2). In Chapter 16 we came across a test that examines whether the population correlation matrix resembles an identity matrix: Bartlett’s test. If the population correlation matrix resembles an identity matrix then it means that every variable correlates very badly with all other variables (i.e. all correlation coefficients are close to zero). If it were an identity matrix then it would mean that all variables are perfectly independent from one another (all correlation coefficients are zero). Given that we are looking for clusters of variables that measure similar things, it should be obvious why this scenario is problematic: if no variables correlate then there are no clusters to find. Bartlett’s test tells us whether our correlation matrix is significantly different from an identity matrix. Therefore, if it is significant then it means that the correlations between variables are (overall) significantly different from zero. So, if Bartlett’s test is significant then it is good news. However, as with any significance test it depends on sample sizes and in factor analysis we typically use very large samples. Therefore, although a nonsignificant Bartlett’s test is certainly cause for concern, a significant test does not necessarily mean that correlations are big enough to make the analysis meaningful. If you do identify any variables, that seem to have very low correlations with lots of other variables, then exclude them from the factor analysis.
The opposite problem is when variables correlate too highly. Although mild multicollinearity is not a problem for factor analysis it is important to avoid extreme multicollinearity (i.e. variables that are very highly correlated) and singularity (variables that are perfectly correlated). As with regression, multicollinearity causes problems in factor analysis because it becomes impossible to determine the unique contribution to a factor of the variables that are highly correlated (as was the case for multiple regression). Multicollinearity does not cause a problem for principal component analysis. Therefore, as well as scanning the correlation matrix for low correlations, we could also look out for very high correlations (r > .8). The problem with a heuristic such as this is that the effect of two variables correlating with r = .9 might be less than the effect of, say, three variables that all correlate at r = .6. In other words, eliminating such highly correlating variables might not be getting at the cause of the multicollinearity (Rockwell, 1975).
Multicollinearity can be detected by looking at the determinant of the R-matrix, denoted |R| (see Jane Superbrain Box 17.3). One simple heuristic is that the determinant of the R-matrix should be greater than 0.00001. However, Haitovsky (1969) proposed a significance test of whether the determinant is zero (i.e. the matrix is singular). If this test is significant it tells us that the correlation matrix is significantly different from a singular matrix, which implies that there is no severe multicollinearity. Simple eh? Well, not quite so simple because SPSS doesn’t do this test! However, it can be done manually without too much trauma:
Haitovsky’s χ²H = [1 + ((2p+5)/6) - N] ln(1 – |R|) (17.6)
in which p is the number of variables in the correlation matrix, N is the total sample size, |R| is the determinant of the correlation matrix and ln is the natural logarithm (this is a standard mathematical function that we came across in Chapter 8 and you can find it on your calculator usually labelled as ln or loge). The resulting test statistic has a chi-square distribution with p(p-1)/2 degrees of freedom. We’ll see how to use this equation in due course.
If you have reason to believe that the correlation matrix has multicollinearity then you could look through the correlation matrix for variables that correlate very highly (R > 0.8) and consider eliminating one of the variables (or more depending on the extent of the problem) before proceeding. You may have to try some trial and error to work out which variables are creating the problem (it’s not always the two with the highest correlation, it could be a larger number of variables with correlations that are not obviously too large).
17.6.2. Rotation
We have already seen that the interpretability of factors can be improved through rotation. Rotation maximizes the loading of each variable on one of the extracted factors while minimizing the loading on all other factors. This process makes it much clearer which variables relate to which factors. Rotation works through changing the absolute values of the variables while keeping their differential values constant. Click on to access the dialog box in Figure 17.9. I’ve discussed the various rotation options in section 17.4.6.1, but, to summarize, the exact choice of rotation will depend on whether or not you think that the underlying factors should be related. If there are theoretical grounds to think that the factors are independent (unrelated) then you should choose one of the orthogonal rotations (I recommend varimax). However, if theory suggests that your factors might correlate then one of the oblique rotations (direct oblimin or promax) should be selected. In this example I’ve selected varimax.
The dialog box also has options for displaying the Rotated solution and a Loading plot. The rotated solution is displayed by default and is essential for interpreting the final rotated analysis. The loading plot will provide a graphical display of each variable plotted against the extracted factors up to a maximum of three factors (unfortunately SPSS cannot produce four- or five-dimensional graphs!). This plot is basically similar to Figure 17.2 and it uses the factor loading of each variable for each factor. With two factors these plots are fairly interpretable, and you should hope to see one group of variables clustered close to the X-axis and a different group of variables clustered around the Y-axis. If all variables are clustered between the axes, then the rotation has been relatively unsuccessful in maximizing the loading of a variable onto a single factor. With three factors these plots can become quite messy and certainly put considerable strain on the visual system! However, they can still be a useful way to determine the underlying structures within the data.
A final option is to set the Maximum Iterations for Convergence, which specifies the number of times that the computer will search for an optimal solution. In most circumstances the default of 25 is more than adequate for SPSS to find a solution for a given data set. However, if you have a large data set (like we have here) then the computer might have difficulty finding a solution (especially for oblique rotation). To allow for the large data set we are using, change the value to 30.
CRAMMING SAM’s Tips Preliminary analysis
• Scan the Correlation Matrix: look for variables that don’t correlate with any other variables, or correlate very highly (r = .9) with one or more other variable. In factor analysis, check that the determinant of this matrix is bigger than 0.00001; if it is then multicollinearity isn’t a problem.
• In the table labelled KMO and Bartlett’s Test the KMO statistic should be greater than 0.5 as a bare minimum; if it isn’t collect more data. Bartlett’s test of sphericity should be significant (the value of Sig. should be less than .05). You can also check the KMO statistic for individual variables by looking at the diagonal of the Anti-Image Matrices: again, these values should be above 0.5 (this is useful for identifying problematic variables if the overall KMO is unsatisfactory).
17.7.2. Factor extraction
The first part of the factor extraction process is to determine the linear components within the data set (the eigenvectors) by calculating the eigenvalues of the R-matrix (see section 17.4.4). We know that there are as many components (eigenvectors) in the R-matrix as there are variables, but most will be unimportant. To determine the importance of a particular vector we look at the magnitude of the associated eigenvalue. We can then apply criteria to determine which factors to retain and which to discard. By default SPSS uses Kaiser’s criterion of retaining factors with eigenvalues greater than 1 (see Figure 17.8).
SPSS Output 17.4 lists the eigenvalues associated with each linear component (factor) before extraction, after extraction and after rotation. Before extraction, SPSS has identified 23 linear components within the data set (we know that there should be as many eigenvectors as there are variables and so there will be as many factors as variables – see section 17.4.4). The eigenvalues associated with each factor represent the variance explained by that particular linear component and SPSS also displays the eigenvalue in terms of the percentage of variance explained (so, factor 1 explains 31.696% of total variance). It should be clear that the first few factors explain relatively large amounts of variance (especially factor 1) whereas subsequent factors explain only small amounts of variance. SPSS then extracts all factors with eigenvalues greater than 1, which leaves us with four factors. The eigenvalues associated with these factors are again displayed (and the percentage of variance explained) in the columns labelled Extraction Sums of Squared Loadings. The values in this part of the table are the same as the values before extraction, except that the values for the discarded factors are ignored (hence, the table is blank after the fourth factor). In the final part of the table (labelled Rotation Sums of Squared Loadings), the eigenvalues of the factors after rotation are displayed. Rotation has the effect of optimizing the factor structure and one consequence for these data is that the relative importance of the four factors is equalized. Before rotation, factor 1 accounted for considerably more variance than the remaining three (31.696% compared to 7.560, 5.725 and 5.336%), but after extraction it accounts for only 16.219% of variance (compared to 14.523, 11.099 and 8.475% respectively).
SPSS Output 17.5 shows the table of communalities before and after extraction. Remember that the communality is the proportion of common variance within a variable (see section 17.4.1). Principal component analysis works on the initial assumption that all variance is common; therefore, before extraction the communalities are all 1 (see column labelled Initial). In effect, all of the variance associated with a variable is assumed to be common variance. Once factors have been extracted, we have a better idea of how much variance is, in reality, common. The communalities in the column labelled Extraction reflect this common variance. So, for example, we can say that 43.5% of the variance associated with question 1 is common, or shared, variance. Another way to look at these communalities is in terms of the proportion of variance explained by the underlying factors. Before extraction, there are as many factors as there are variables, so all variance is explained by the factors and communalities are all 1. However, after extraction some of the factors are discarded and so some information is lost. The retained factors cannot explain all of the variance present in the data, but they can explain some. The amount of variance in each variable that can be explained by the retained factors is represented by the communalities after extraction.
SPSS Output 17.5 also shows the component matrix before rotation. This matrix contains the loadings of each variable onto each factor. By default SPSS displays all loadings; however, we requested that all loadings less than 0.4 be suppressed in the output (see Figure 17.11) and so there are blank spaces for many of the loadings. This matrix is not particularly important for interpretation, but it is interesting to note that before rotation most variables load highly onto the first factor (that is why this factor accounts for most of the variance in SPSS Output 17.4).
At this stage SPSS has extracted four factors. Factor analysis is an exploratory tool and so it should be used to guide the researcher to make various decisions: you shouldn’t leave the computer to make them. One important decision is the number of factors to extract. In section 17.4.5 we saw various criteria for assessing the importance of factors. By Kaiser’s criterion we should extract four factors and this is what SPSS has done. However, this criterion is accurate when there are less than 30 variables and communalities after extraction are greater than 0.7 or when the sample size exceeds 250 and the average communality is greater than 0.6. The communalities are shown in SPSS Output 17.5, and only one exceeds 0.7. The average of the communalities can be found by adding them up and dividing by the number of communalities (11.573/23 = 0.503). So, on both grounds Kaiser’s rule may not be accurate. However, you should consider the huge sample that we have, because the research into Kaiser’s criterion gives recommendations for much smaller samples. By Jolliffe’s criterion (retain factors with eigenvalues greater than 0.7) we should retain 10 factors (see SPSS Output 17.4), but there is little to recommend this criterion over Kaiser’s. As a final guide we can use the scree plot which we asked SPSS to produce by using the option in Figure 17.8. The scree plot is shown in SPSS Output 17.6. This curve is difficult to interpret because it begins to tail off after three factors, but there is another drop after four factors before a stable plateau is reached. Therefore, we could probably justify retaining either two or four factors. Given the large sample, it is probably safe to assume Kaiser’s criterion; however, you might like to rerun the analysis specifying that SPSS extract only two factors (see Figure 17.8) and compare the results.
SPSS Output 17.7 shows an edited version of the reproduced correlation matrix that was requested using the option in Figure 17.7. The top half of this matrix (labelled Reproduced Correlations) contains the correlation coefficients between all of the questions based on the factor model. The diagonal of this matrix contains the communalities after extraction for each variable (you can check the values against SPSS Output 17.5).
The correlations in the reproduced matrix differ from those in the R-matrix because they stem from the model rather than the observed data. If the model were a perfect fit of the data then we would expect the reproduced correlation coefficients to be the same as the original correlation coefficients. Therefore, to assess the fit of the model we can look at the differences between the observed correlations and the correlations based on the model. For example, if we take the correlation between questions 1 and 2, the correlation based on the observed data is −0.099 (taken from SPSS Output 17.1). The correlation based on the model is −0.112, which is slightly higher. We can calculate the difference as follows:
residual = robserved − rfrom model
residualQ1Q2 = (−0.099) − (−0.112)
= 0.013
You should notice that this difference is the value quoted in the lower half of the reproduced matrix (labelled Residual) for questions 1 and 2. Therefore, the lower half of the reproduced matrix contains the differences between the observed correlation coefficients and the ones predicted from the model. For a good model these values will all be small. In fact, we want most values to be less than 0.05. Rather than scan this huge matrix, SPSS provides a footnote summary, which states how many residuals have an absolute value greater than 0.05. For these data there are 91 residuals (35%) that are greater than 0.05. There are no hard and fast rules about what proportion of residuals should be below 0.05; however, if more than 50% are greater than 0.05 you probably have grounds for concern.
CRAMMING SAM’s Tips Factor extraction
• To decide how many factors to extract look at the table labelled Communalities and the column labelled Extraction. If these values are all 0.7 or above and you have less than 30 variables then the SPSS default option for extracting factors is fine (Kaiser’s criterion of retaining factors with eigenvalues greater than 1). Likewise, if your sample size exceeds 250 and the average of the communalities is 0.6 or greater then the default option is fine. Alternatively, with 200 or more participants the scree plot can be used.
• Check the bottom of the table labelled Reproduced Correlations for the percentage of ‘nonredundant residuals with absolute values > 0.05’. This percentage should be less than 50% and the smaller it is, the better.
17.9. Reliability analysis
17.9.1. Measures of reliability
If you’re using factor analysis to validate a questionnaire, it is useful to check the reliability of your scale.
Reliability means that a measure (or in this case questionnaire) should consistently reflect the construct that it is measuring. One way to think of this is that, other things being equal, a person should get the same score on a questionnaire if they complete it at two different points in time (we have already discovered that this is called test–retest reliability). So, someone who is terrified of statistics and who scores highly on our SAQ should score similarly highly if we tested them a month later (assuming they hadn’t gone into some kind of statistics-anxiety therapy in that month). Another way to look at reliability is to say that two people who are the same in terms of the construct being measured should get the same score. So, if we took two people who were equally statistics-phobic, then they should get more or less identical scores on the SAQ. Likewise, if we took two people who loved statistics, they should both get equally low scores. It should be apparent that if we took someone who loved statistics and someone who was terrified of it, and they got the same score on our questionnaire, then it wouldn’t be an accurate measure of statistical anxiety! In statistical terms, the usual way to look at reliability is based on the idea that individual items (or sets of items) should produce results consistent with the overall questionnaire. So, if we take someone scared of statistics, then their overall score on the SAQ will be high; if the SAQ is reliable then if we randomly select some items from it the person’s score on those items should also be high.
The simplest way to do this in practice is to use split-half reliability. This method randomly splits the data set into two. A score for each participant is then calculated based on each half of the scale. If a scale is very reliable a person’s score on one half of the scale should be the same (or similar) to their score on the other half: therefore, across several participants, scores from the two halves of the questionnaire should correlate perfectly (well, very highly). The correlation between the two halves is the statistic computed in the split-half method, with large correlations being a sign of reliability. The problem with this method is that there are several ways in which a set of data can be split into two and so the results could be a product of the way in which the data were split. To overcome this problem, Cronbach (1951) came up with a measure that is loosely equivalent to splitting data in two in every possible way and computing the correlation coefficient for each split. The average of these values is equivalent to Cronbach’s alpha, α, which is the most common measure of scale reliability. [9]
Cronbach’s α is:
which may look complicated, but actually isn’t. The first thing to note is that for each item on our scale we can calculate two things: the variance within the item, and the covariance between a particular item and any other item on the scale. Put another way, we can construct a variance–covariance matrix of all items. In this matrix the diagonal elements will be the variance within a particular item, and the off-diagonal elements will be covariances between pairs of items. The top half of the equation is simply the number of items (N) squared multiplied by the average covariance between items (the average of the off-diagonal elements in the aforementioned variance–covariance matrix). The bottom half is just the sum of all the item variances and item covariances (i.e. the sum of everything in the variance–covariance matrix).
There is a standardized version of the coefficient too, which essentially uses the same equation except that correlations are used rather than covariances, and the bottom half of the equation uses the sum of the elements in the correlation matrix of items (including the ones that appear on the diagonal of that matrix). The normal alpha is appropriate when items on a scale are summed to produce a single score for that scale (the standardized alpha is not appropriate in these cases). The standardized alpha is useful, though, when items on a scale are standardized before being summed.
[9] Although this is the easiest way to conceptualize Cronbach’s α, whether or not it is exactly equal to the average of all possible split-half reliabilities depends on exactly how you calculate the split-half reliability (see the glossary for computational details). If you use the Spearman–Brown formula, which takes no account of item standard deviations, then Cronbach’s α will be equal to the average split half-reliability only when the item standard deviations are equal; otherwise α will be smaller than the average. However, if you use a formula for split-half reliability that does account for item standard deviations (such as Flanagen, 1937; Rulon, 1939) then α will always equal the average split-half reliability (see Cortina, 1993).